Examples¶
Here is a collection of grid examples and how to write them to GGD.
Example 1: Minimalistic unstructured grid example¶
In this example we will describe a simple unstructured grid (single time slice) consisting of one 2D object - surface to present the basic concepts behind GGD.
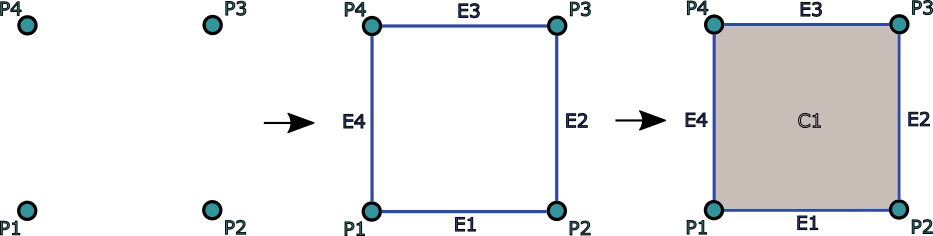
Grid consisting of one 2D object which is composed by 4 points / 4 edges / 1 2D cell.¶
Defining the grid¶
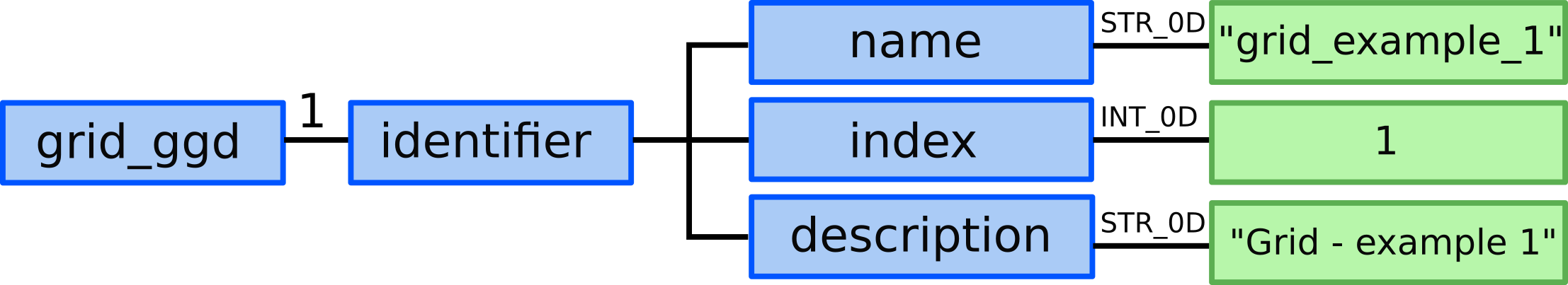
1.) We need to allocate one grid_ggd AOS (one time slice).
2.) In grid_ggd(1).identifier we define the grid name, index and
description:
grid_ggd(1).identifier.name = "grid_example_1"grid_ggd(1).identifier.index = 1grid_ggd(1).identifier.description = "Grid - example 1"
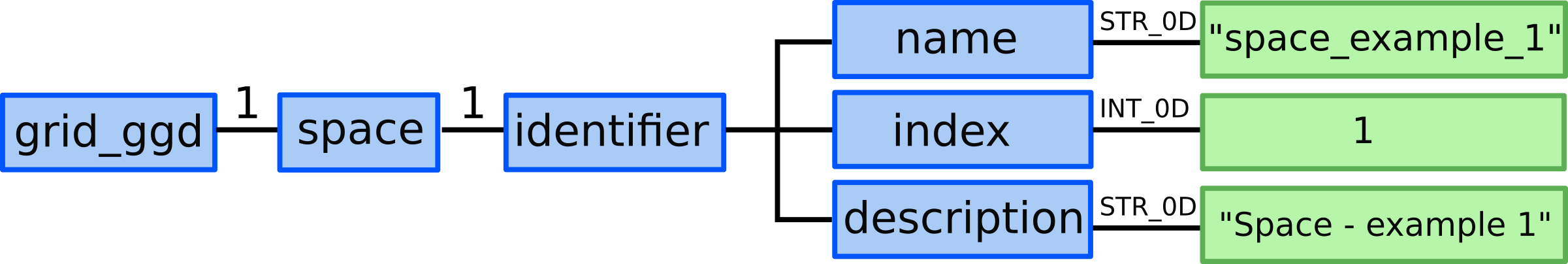
3.) We allocate one grid_ggd(1).space AOS.
4.) In grid_ggd(1).space(1).identifier node we define the space name,
index and description:
grid_ggd(1).space(1).identifier.name = "space_example_1"grid_ggd(1).space(1).identifier.index = 1grid_ggd(1).space(1).identifier.description = "Space - example 1"
5.) In grid_ggd(1).space(1).geometry_type.index leaf we define
standard geometry -> index 0
grid_ggd(1).space(1).geometry_type.index = 0

6.) In grid_ggd(1).space(1).coordinates_type leaf we define two coordinates
following the coordinate_identifier this way defining 2D space. We
define 1 and 2 meaning X and Y coordinates:
grid_ggd(1).space(1).coordinate_types = [1, 2]

7.) We allocate three grid_ggd(1).space(1).objects_per_dimension AOSs:
for 0D objects - points, 1D objects - edges/lines, and
2D objects - surfaces/2D cells.
8.1) We allocate 4 ...objects_per_dimension(1).object AOS as our grid
consists of 4 points.
8.2) We define 4 points by defining their geometry (note that the coordinates are simplified according to the example):
...objects_per_dimension(1).object(1).geometry = [0.0, 0.0]...objects_per_dimension(1).object(2).geometry = [1.0, 0.0]...objects_per_dimension(1).object(3).geometry = [1.0, 1.0]...objects_per_dimension(1).object(4).geometry = [0.0, 1.0]
9.1) We allocate 4 ...objects_per_dimension(2).object AOS as our grid
consists of 4 lines.
9.2) We define 4 lines by defining nodes that compose each line (see figure Grid consisting of one 2D object which is composed by 4 points / 4 edges / 1 2D cell.):
...objects_per_dimension(2).object(1).nodes = [1, 2]...objects_per_dimension(2).object(2).nodes = [2, 3]...objects_per_dimension(2).object(3).nodes = [3, 4]...objects_per_dimension(2).object(4).nodes = [4, 1]
10.1) We allocate 1 ...objects_per_dimension(3).object AOS as our grid
consists of 1 quadrilateral cell.
10.2) We define 1 quadrilateral cell by defining 4 nodes that compose the quadrilateral (see figure Grid consisting of one 2D object which is composed by 4 points / 4 edges / 1 2D cell.):
...objects_per_dimension(3).object(1).nodes = [1, 2, 3, 4]
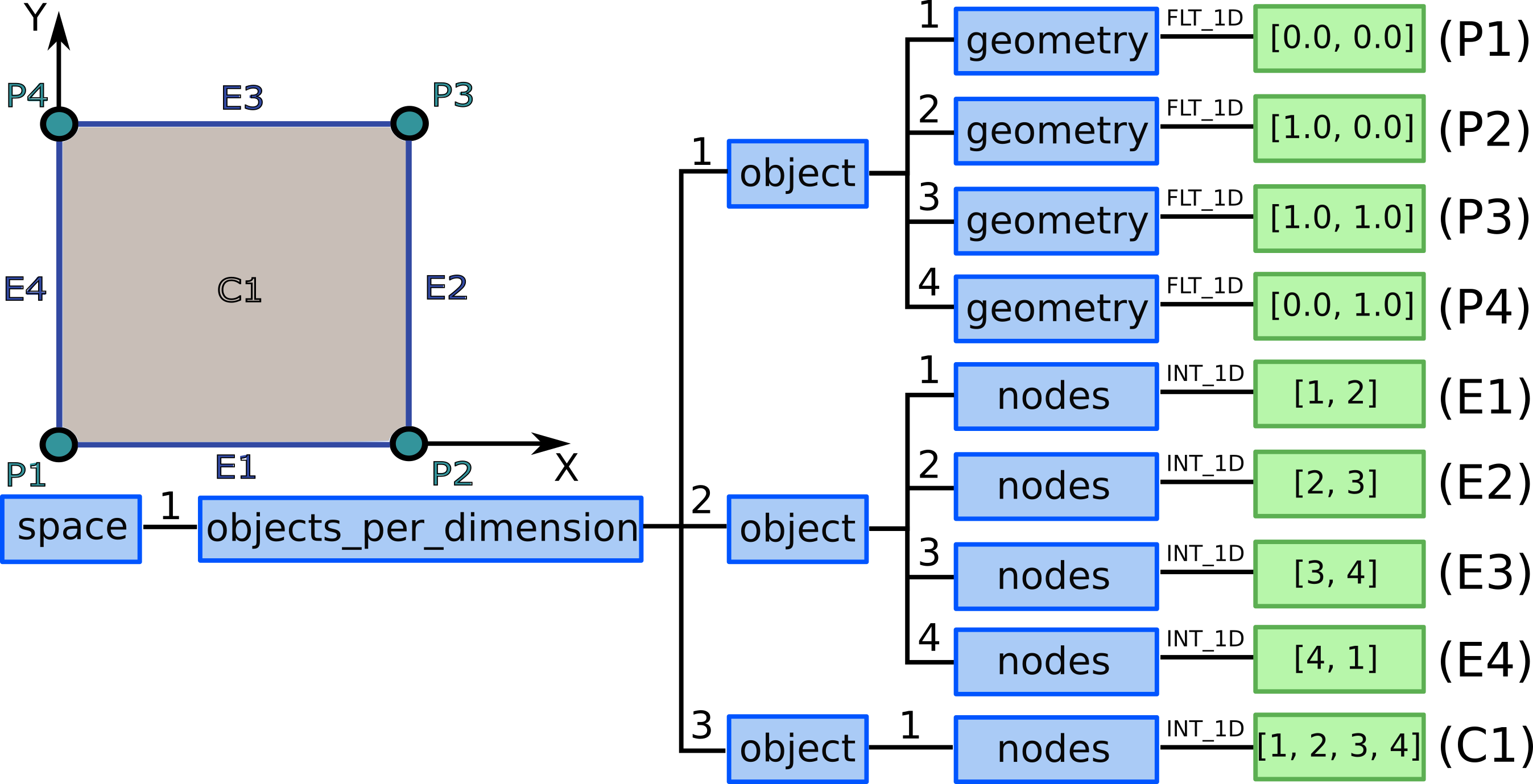
Now we have all grid objects in the domain defined.
Defining grid subsets¶
In our case we don’t have any specific grid subsets, however, we still need to define the grid subsets correlating to the 0D, 1D and 2D objects in the domain. This is due to Data Dictionary description, as physical quantities correlate directly to grid subsets and not to the grid itself.
The agreed grid subset labels can be found here:
ggd_subset_identifier.
11.1) Allocate 3 grid_ggd(1).grid_subset AOSs (nodes, faces, 2D cells):
11.2) Define nodes grid subset. By definition nodes grid subset is composed by all 0D objects defined in this grid_ggd AOS so the element AOS can be left empty.
grid_ggd(1).grid_subset(1).dimension = 1grid_ggd(1).grid_subset(1).identifier.name = "nodes"grid_ggd(1).grid_subset(1).identifier.index = 1grid_ggd(1).grid_subset(1).identifier.description = "All nodes in the domain"
11.3) Define faces grid subset. By definition faces grid subset is composed by all 0D objects defined in this grid_ggd AOS so the element AOS can be left empty.
grid_ggd(1).grid_subset(2).dimension = 2grid_ggd(1).grid_subset(2).identifier.name = "faces"grid_ggd(1).grid_subset(2).identifier.index = 2grid_ggd(1).grid_subset(2).identifier.description = "All lines in the domain"
11.4) Define cells grid subset. By definition cells grid subset is composed by all 0D objects defined in this grid_ggd AOS so the element AOS can be left empty.
grid_ggd(1).grid_subset(3).dimension = 3grid_ggd(1).grid_subset(3).identifier.name = "cells"grid_ggd(1).grid_subset(3).identifier.index = 5grid_ggd(1).grid_subset(3).identifier.description = "All 2D cells in the domain"
With this last step the grid description of our minimalistic unstructured grid example is finished.
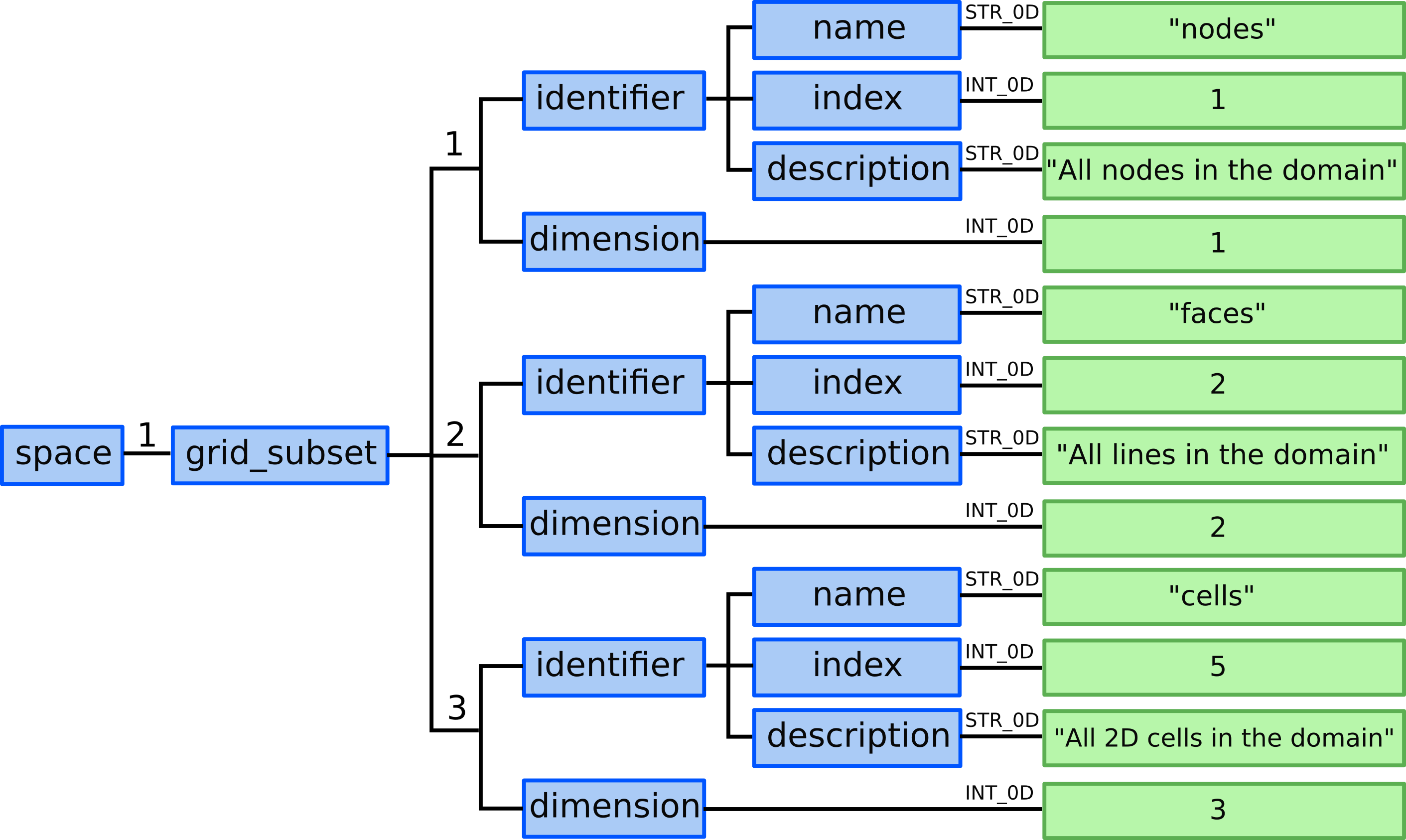
Defining the three base grid subsets - nodes, faces and cells. Note that the dimension ID doesn’t start with 0 as 0 values are in Data Dictionary reserved for undefined indices.¶
Defining physical quantities¶
Suppose that we have electron temperature and ion specie Ne+2 density
quantities/values for single time
slice at time 1.0ms that correlate to our 4 nodes and to our one
2D cell. Defining them in GGD is done in the next way:
12.) We need to allocate one ggd AOS (one time slice).
13.) We specify time of this time slice:
ggd(1).time= 1.0
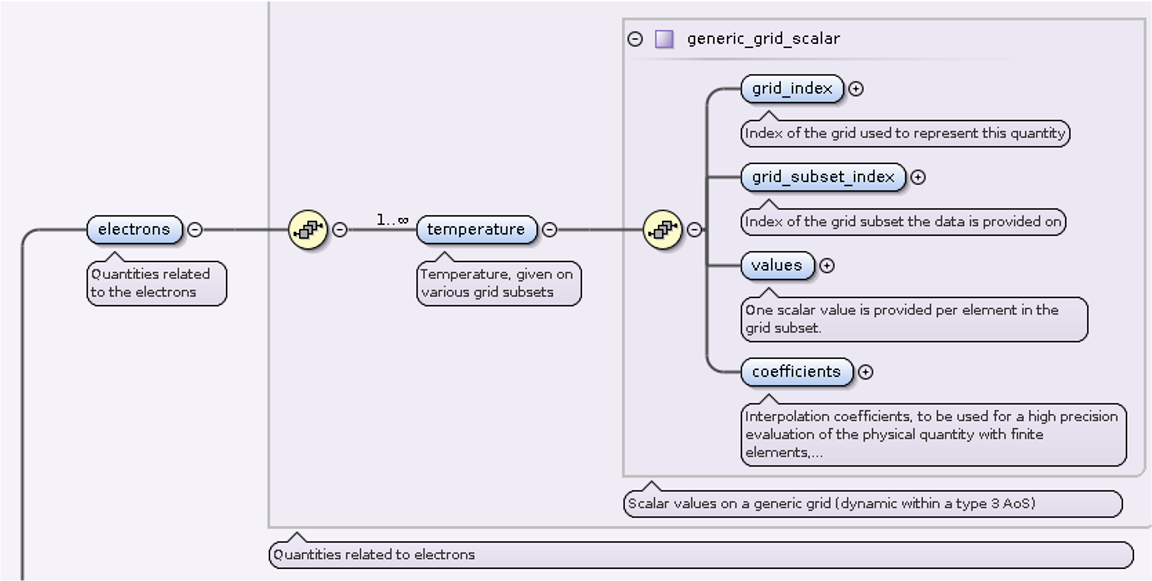
Electron temperature¶
14.) We allocate two ggd(1).electrons.temperature AOS as in this case
we have available electron temperature quantities for two grid subsets.
15.) In ggd(1).electrons.temperature(1) we define grid_index,
grid_subset_index and values leafs.
grid_index and grid_subset_index are used to navigate
to grid which has grid_ggd(:).identifier.index == grid_index
and :grid_ggd(:).grid_subset(:).identifier.index == grid_subset_index.
In this case it would navigate to our nodes grid subset.
ggd(1).electrons.temperature(1).grid_index= 1ggd(1).electrons.temperature(1).grid_subset_index= 1ggd(1).electrons.temperature(1).values= e.g. [1.1, 2.2, 3.3, 4.4] (one value per point)
16.) In ggd(1).electrons.temperature(2) we define grid_index,
grid_subset_index and values leafs.
grid_index and grid_subset_index are used to navigate
to grid which has grid_ggd(:).identifier.index == grid_index
and :grid_ggd(:).grid_subset(:).identifier.index == grid_subset_index.
In this case it would navigate to our cells grid subset.
ggd(1).electrons.temperature(2).grid_index= 1ggd(1).electrons.temperature(2).grid_subset_index= 2ggd(1).electrons.temperature(2).values= e.g. [2.5] (one value per cell)
Ion density - specie Ne+2¶
17.) We allocate one ggd(1).ion AOS (as we have one ion specie).
18.) We define ion specie label:
ggd(1).ion(1).label= “Ne+”
19.) We allocate one ion state ggd(1).ion(1).state AOS as we have one
state - Ne+2. Other states could be Ne+3 ,**Ne+4**, etc., however we
don’t have those in this example.
20.) We allocate two ggd(1).ion(1).state(1).density AOS (for nodes and
cells grid subsets).
21.) We define ion specie state label:
ggd(1).ion(1).label= “Ne+2”
22.) In ggd(1).ion(1).state(1).density(1) we define grid_index,
grid_subset_index and values leafs.
grid_index and grid_subset_index are used to navigate
to grid which has grid_ggd(:).identifier.index == grid_index
and :grid_ggd(:).grid_subset(:).identifier.index == grid_subset_index.
In this case it would navigate to our nodes grid subset.
ggd(1).ion(1).state(1).density(1).grid_index= 1ggd(1).ion(1).state(1).density(1).grid_subset_index= 1ggd(1).ion(1).state(1).density(1).values= e.g. [0.1, 0.2, 0.3, 0.4] (one value per point)
23.) In ggd(1).ion(1).state(1).density(2) we define grid_index,
grid_subset_index and values leafs.
grid_index and grid_subset_index are used to navigate
to grid which has grid_ggd(:).identifier.index == grid_index
and :grid_ggd(:).grid_subset(:).identifier.index == grid_subset_index.
In this case it would navigate to our cells grid subset.
ggd(1).ion(1).state(1).density(2).grid_index= 1ggd(1).ion(1).state(1).density(2).grid_subset_index= 2ggd(1).ion(1).state(1).density(2).values= e.g. [0.3] (one value per cell)
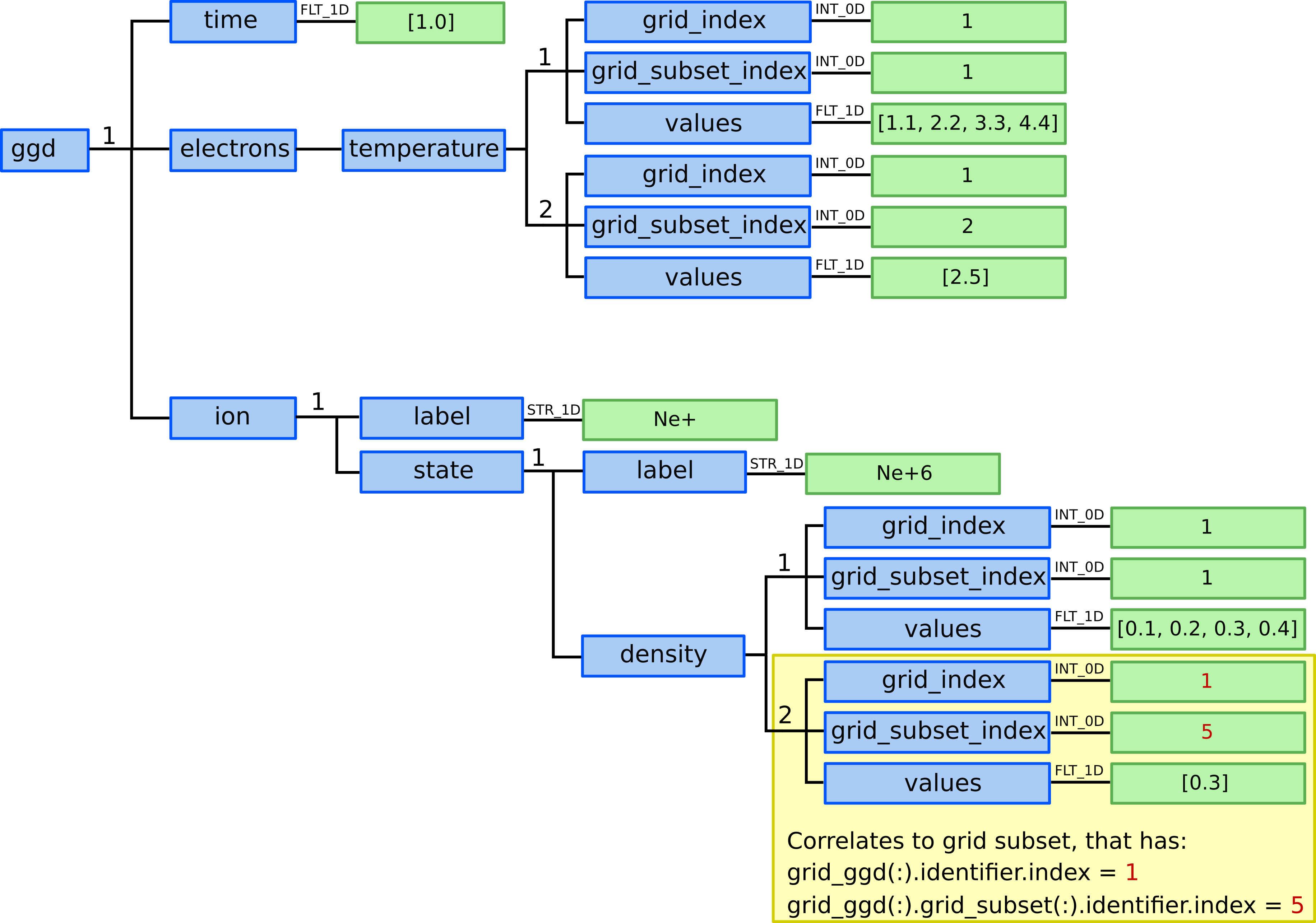
Defining physical quantities and their correlation to grid subsets.¶
Example 2: Minimalistic structured grid example¶
In this example we will describe a simple structured grid (two time slices) consisting of 5 points in X direction, and 3 points in Y direction to present the basic concepts behind GGD.
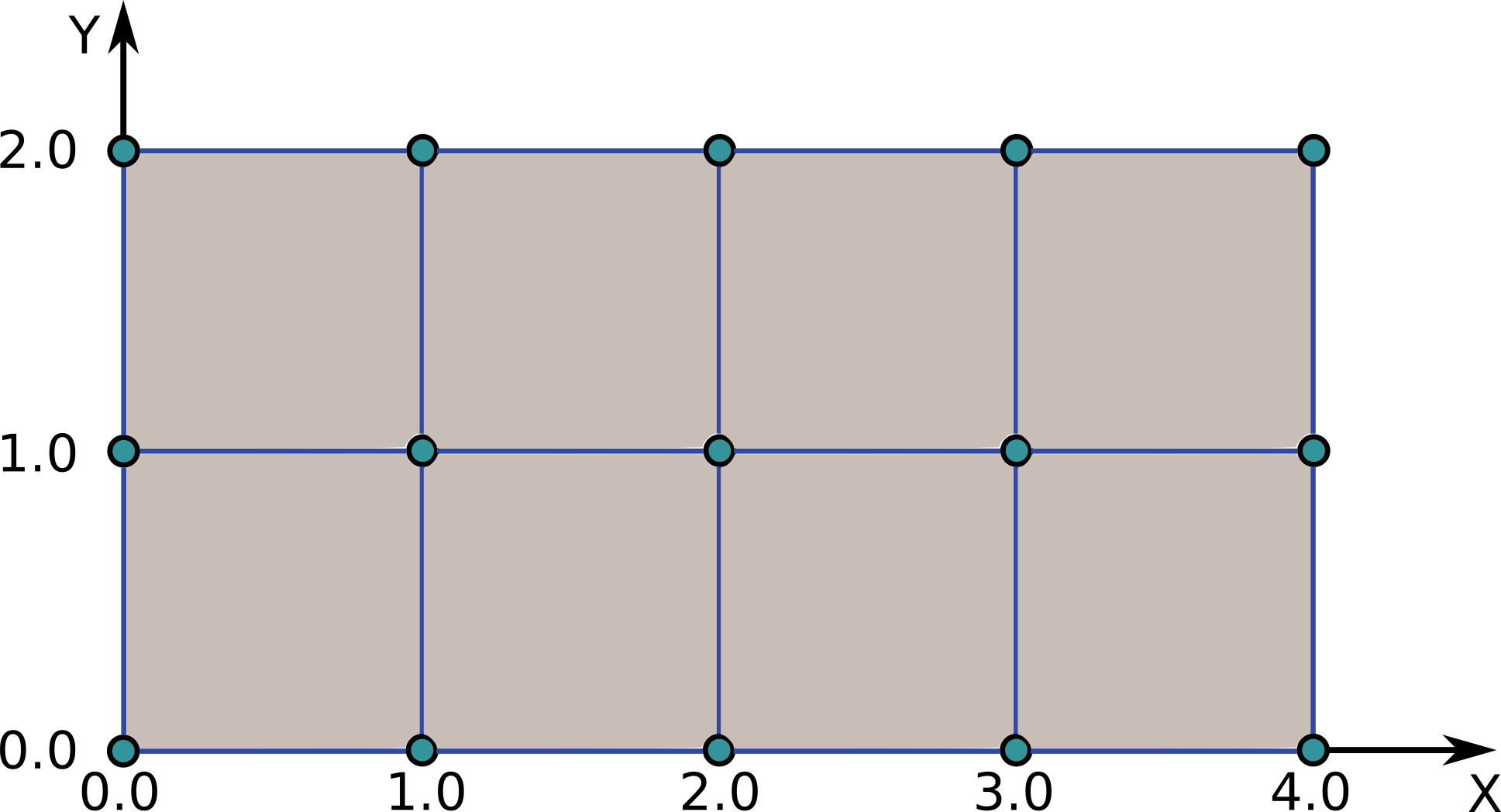
Grid consisting of consisting of 5 points in X direction, and 3 points in Y direction.¶
Defining the grid¶
1.) We need to allocate one grid_ggd AOS. Grid will remain static through the next time slice so we can define it only for one “time slice”.
2.) In grid_ggd(1).identifier we define the grid name, index and
description:
grid_ggd(1).identifier.name = "grid_example_2"grid_ggd(1).identifier.index = 1grid_ggd(1).identifier.description = "Grid - example 2"
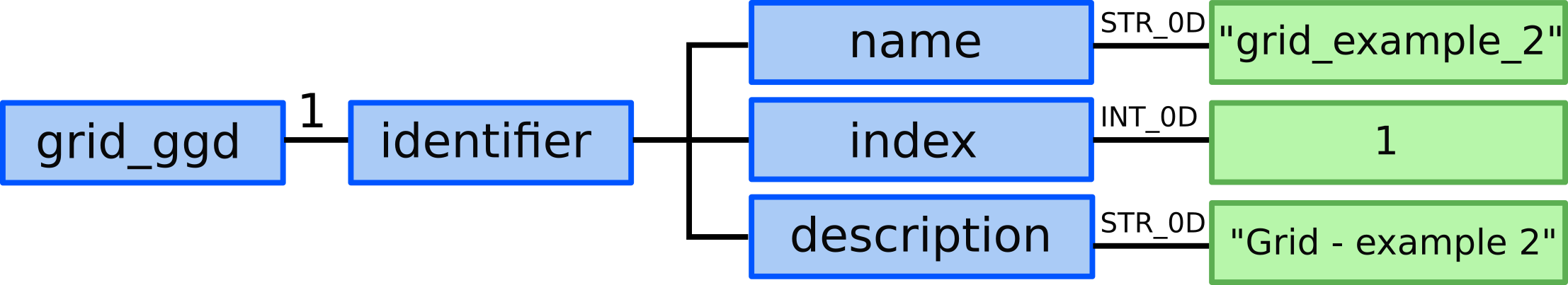
3.) We allocate two grid_ggd(1).space AOSs. One for X direction and
the second for Y direction.
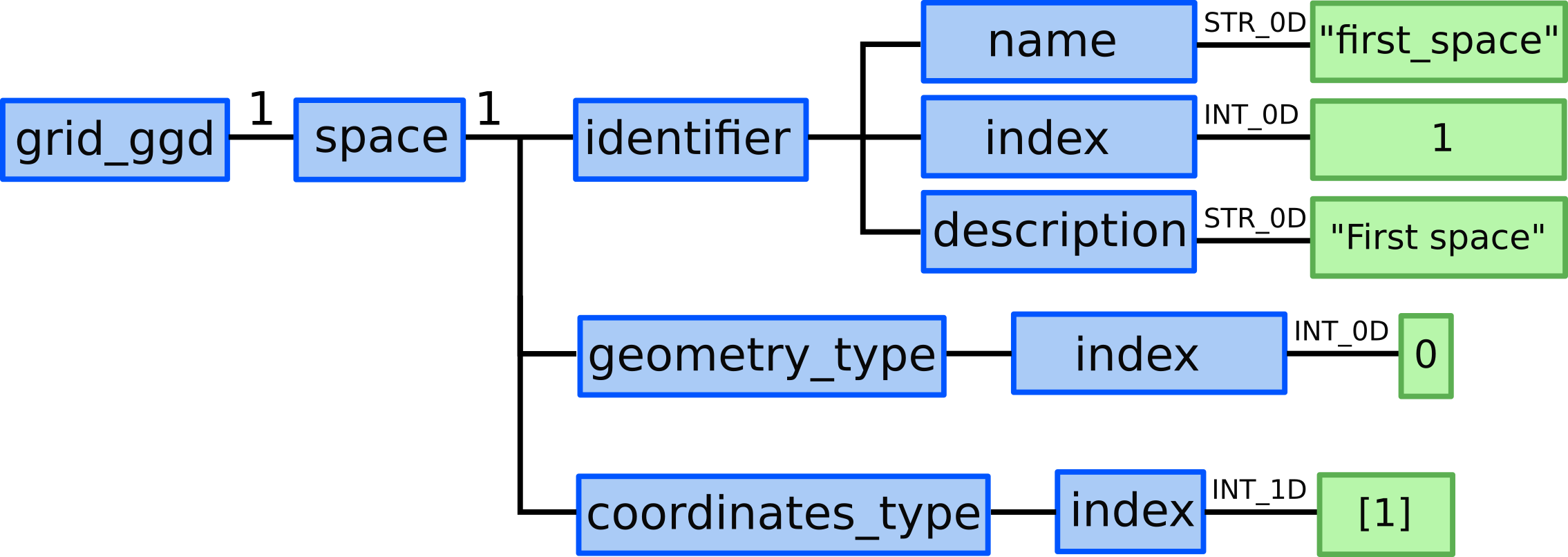
Space 1 - X direction¶
4.) In grid_ggd(1).space(1).identifier node we define the space name,
index and description:
grid_ggd(1).space(1).identifier.name = "first_space"grid_ggd(1).space(1).identifier.index = 1grid_ggd(1).space(1).identifier.description = "First Space"
5.) In grid_ggd(1).space(1).geometry_type.index leaf we define
standard geometry -> index 0.
6.) In grid_ggd(1).space(1).coordinates_type leaf we define one
coordinate following the coordinate_identifier this way defining 1D space. We define 1 meaning X coordinate:
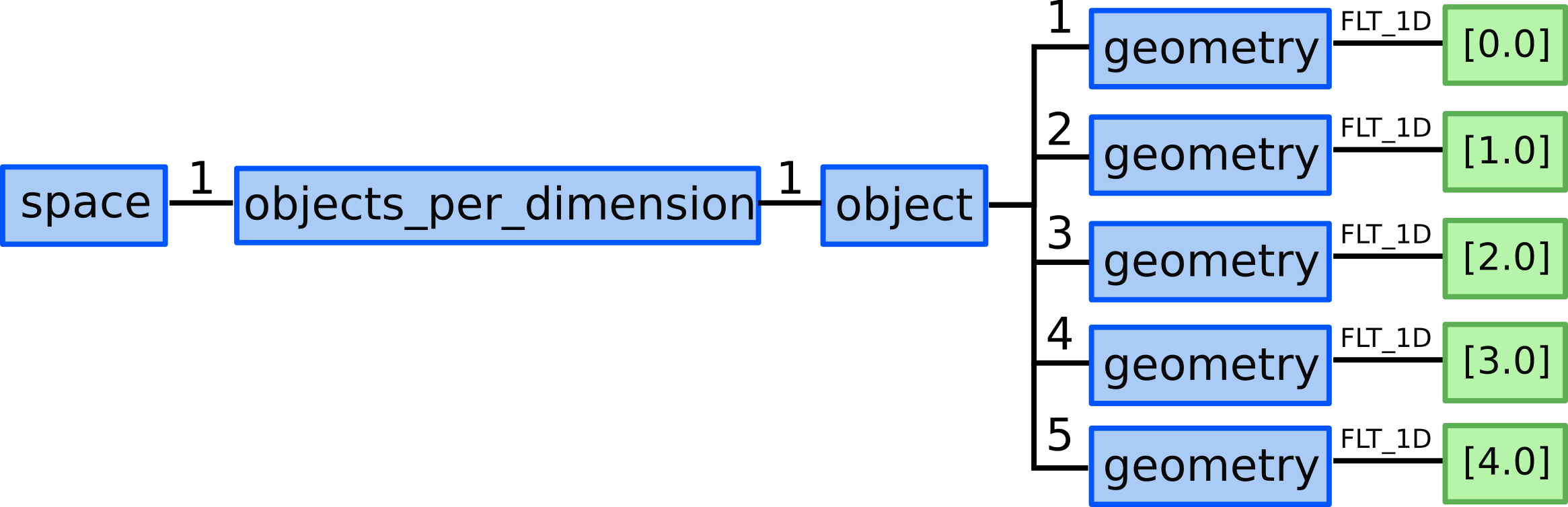
7.) We allocate one grid_ggd(1).space(1).objects_per_dimension AOSs:
for 0D objects - points in X direction.
8.1) We allocate 5 ...objects_per_dimension(1).object AOS as our
structured grid consists of 5 points in X direction.
8.2) We define 5 points by defining their geometry:
...objects_per_dimension(1).object(1).geometry = [0.0]...objects_per_dimension(1).object(2).geometry = [1.0]...objects_per_dimension(1).object(3).geometry = [2.0]...objects_per_dimension(1).object(4).geometry = [3.0]...objects_per_dimension(1).object(5).geometry = [4.0]
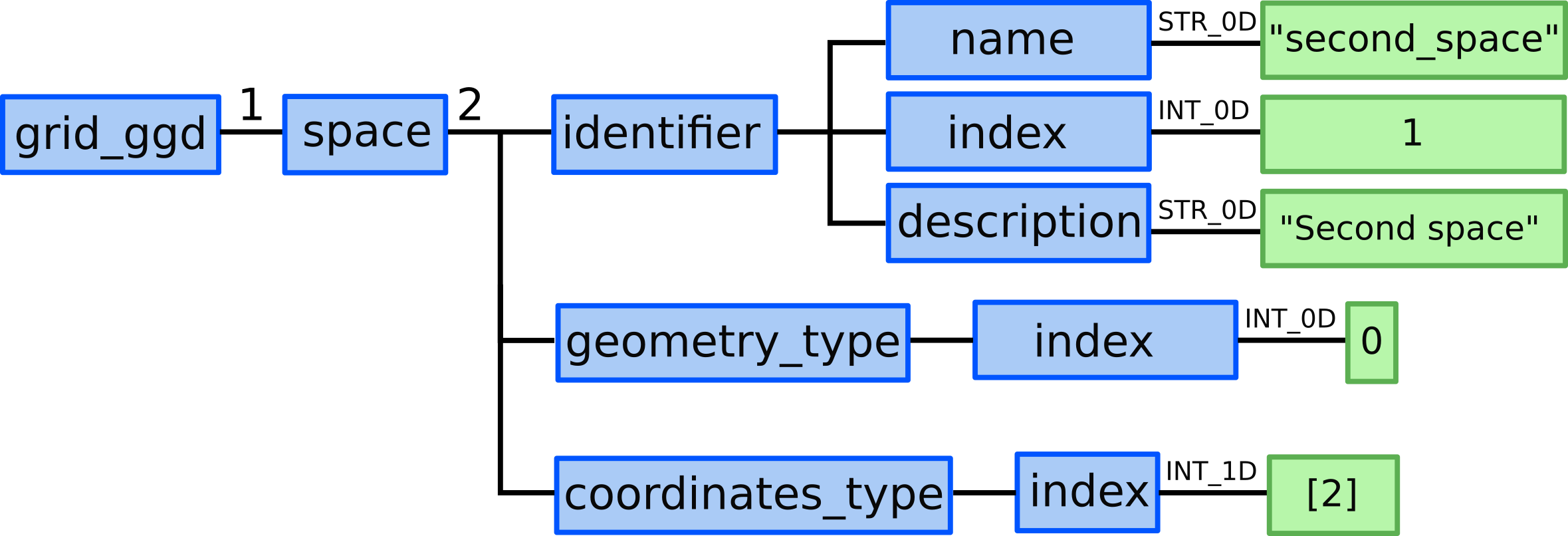
Space 2 - Y direction¶
9.) In grid_ggd(1).space(2).identifier node we define the space name,
index and description:
grid_ggd(1).space(2).identifier.name = "second_space"grid_ggd(1).space(2).identifier.index = 1grid_ggd(1).space(2).identifier.description = "Second Space"
10.) In grid_ggd(1).space(2).geometry_type.index leaf we define
standard geometry -> index 0
11.) In grid_ggd(1).space(2).coordinates_type leaf we define one
coordinate following the coordinate_identifier this way defining 1D space. We define 2 meaning Y coordinate:
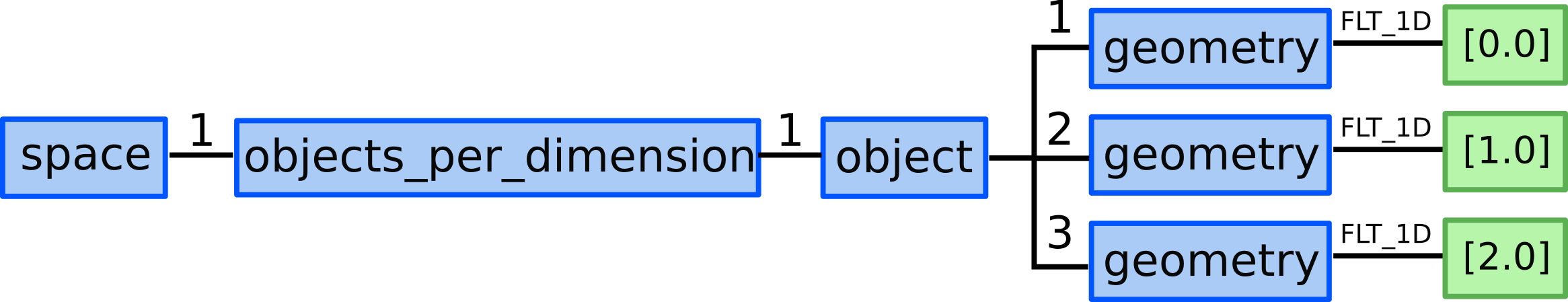
12.) We allocate one grid_ggd(1).space(2).objects_per_dimension AOSs:
for 0D objects - points in X direction.
13.1) We allocate 3 ...objects_per_dimension(1).object AOS as our
structured grid consists of 3 points in Y direction.
13.2) We define 5 points by defining their geometry:
...objects_per_dimension(1).object(1).geometry = [0.0]...objects_per_dimension(1).object(2).geometry = [1.0]...objects_per_dimension(1).object(3).geometry = [2.0]
Defining grid subsets¶
We need to define the grid subsets correlating to the 0D, 1D and 2D objects in the domain. This is due to Data Dictionary description, as physical quantities correlate directly to grid subsets and to the grid itself.
The agreed grid subset labels can be found here:
ggd_subset_identifier.
14.2) Allocate 3 grid_subsets (nodes, faces, 2D cells):
Base grid subsets¶
15.2) Define nodes grid subset. By definition nodes grid subset is composed by all 0D objects defined in this grid_ggd AOS so the element AOS can be left empty.
grid_ggd(1).grid_subset(1).dimension = 1grid_ggd(1).grid_subset(1).identifier.name = "nodes"grid_ggd(1).grid_subset(1).identifier.index = 1grid_ggd(1).grid_subset(1).identifier.description = "All nodes in the domain"
15.3) Define faces grid subset. By definition faces grid subset is composed by all 0D objects defined in this grid_ggd AOS so the element AOS can be left empty.
grid_ggd(1).grid_subset(2).dimension = 2grid_ggd(1).grid_subset(2).identifier.name = "faces"grid_ggd(1).grid_subset(2).identifier.index = 2grid_ggd(1).grid_subset(2).identifier.description = "All lines in the domain"
15.4) Define cells grid subset. By definition cells grid subset is composed by all 0D objects defined in this grid_ggd AOS so the element AOS can be left empty.
grid_ggd(1).grid_subset(3).dimension = 3grid_ggd(1).grid_subset(3).identifier.name = "cells"grid_ggd(1).grid_subset(3).identifier.index = 3grid_ggd(1).grid_subset(3).identifier.description = "All 2D cells in the domain"

Defining three base grid subsets.¶
Defining physical quantities¶
Suppose that we have electron temperature quantities/values for
two time slices at times 1.0ms and 2.0ms that correlate to our
15 points (we assume that we don’t have any data for 2D cells). Defining
them in GGD is done in the next way:
16.) We need to allocate two ggd AOS (two time slices).
17.) We specify time value for both time slices:
ggd(1).time= 1.0ggd(2).time= 2.0
Electron temperature - First time slice¶
18.) We allocate one ggd(1).electrons.temperature AOS as in this case
we have available electron temperature quantities for one grid subset.
19.) In ggd(1).electrons.temperature(1) we define grid_index,
grid_subset_index and values leafs.
grid_index and grid_subset_index are used to navigate
to grid which has grid_ggd(:).identifier.index == grid_index
and :grid_ggd(:).grid_subset(:).identifier.index == grid_subset_index.
In this case it would navigate to our nodes grid subset.
ggd(1).electrons.temperature(1).grid_index= 1ggd(1).electrons.temperature(1).grid_subset_index= 1ggd(1).electrons.temperature(1).values= e.g. [1.1, 2.2, …, 15.15] (one value per point, 15 values)
Electron temperature - Second time slice¶
20.) We allocate one ggd(1).electrons.temperature AOS as in this case
we have available electron temperature quantities for one grid subset.
21.) In ggd(1).electrons.temperature(1) we define grid_index,
grid_subset_index and values leafs.
grid_index and grid_subset_index are used to navigate
to grid which has grid_ggd(:).identifier.index == grid_index
and :grid_ggd(:).grid_subset(:).identifier.index == grid_subset_index.
In this case it would navigate to our nodes grid subset.
ggd(2).electrons.temperature(1).grid_index= 1ggd(2).electrons.temperature(1).grid_subset_index= 1ggd(2).electrons.temperature(1).values= e.g. [2.2, 3.3, …, 16.16] (one value per point, 15 values)
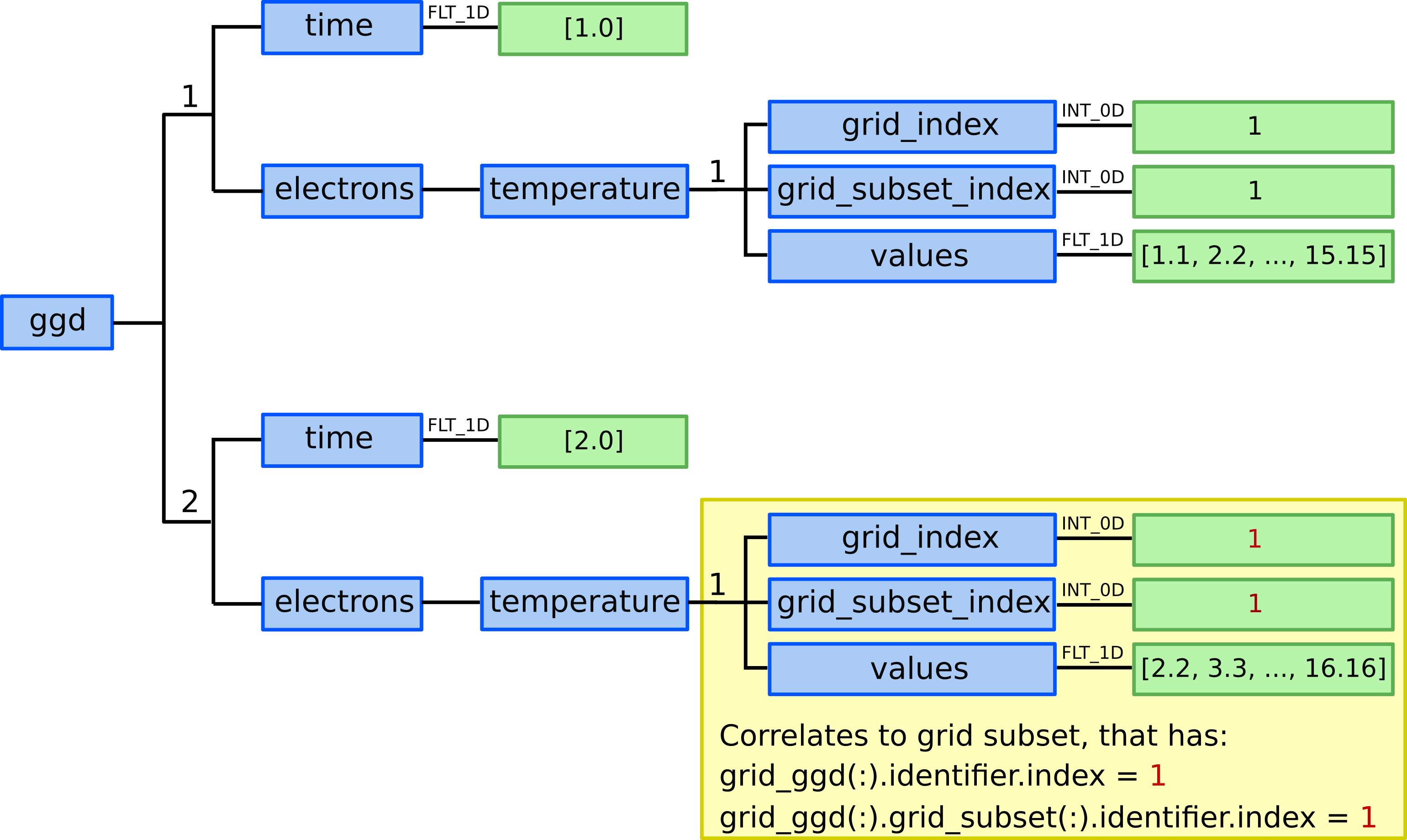
Defining electron temperature data fields for two time slices.¶
Example 3: Structured grid subset example¶
Following the Example 2: Minimalistic structured grid example, lets assume that the two right-most 2D cells represent our outer divertor grid subset, just for purposes to explain the grid subset concepts when dealing with structured grids.
Outer divertor grid subset¶
Our 4-th grid subset represents outer divertor region (just for explanation purposes), as seen in the figure below.
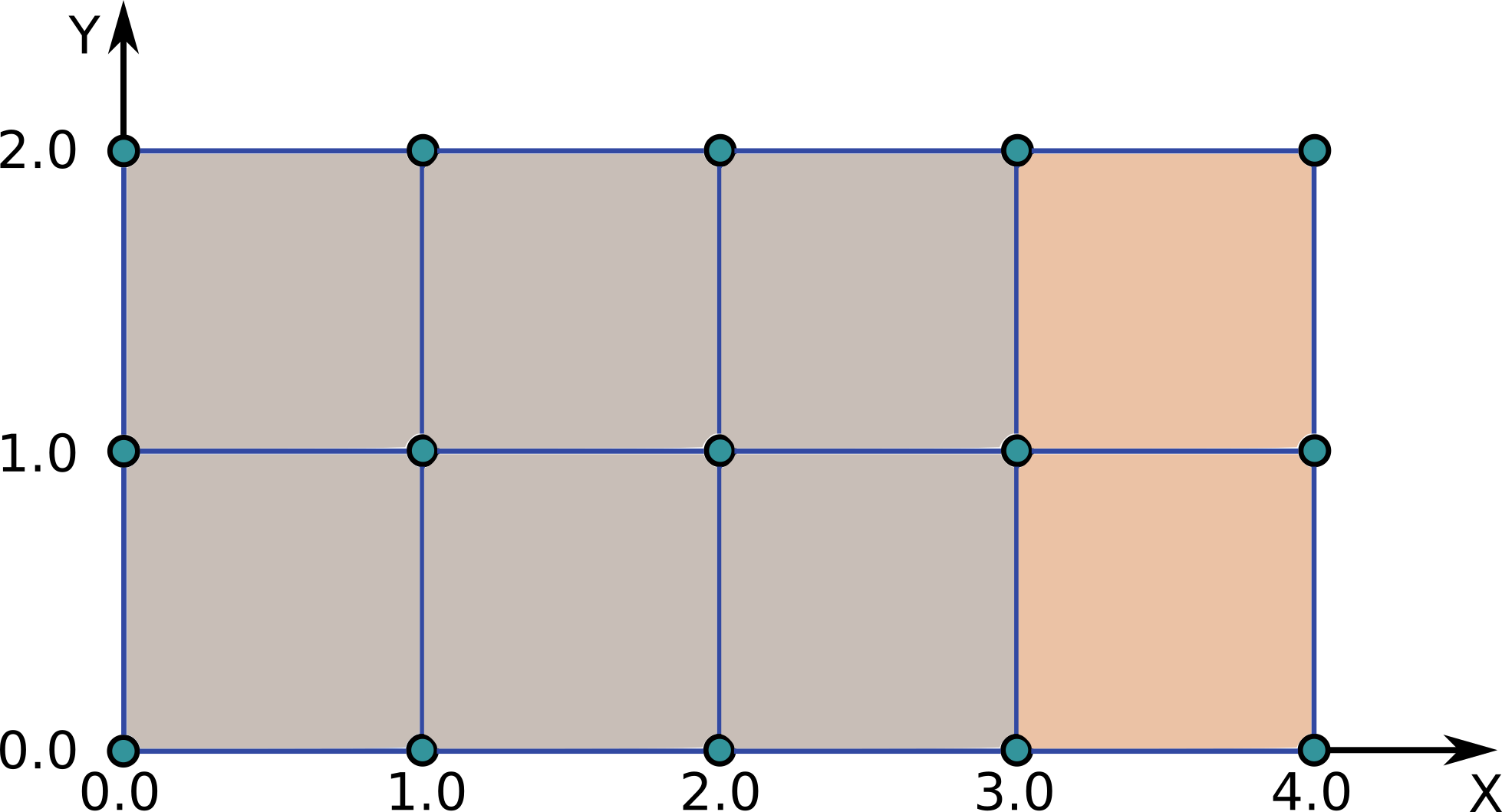
Now we need to define the elements which compose this grid subset. In this case, the elemets are 2D cells, and those 2D cells are composed by objects, in this case points defined on 1D spaces. So we will have 2 elements, each composed by 2x4 points which are defined on 1D spaces.
1.) Describe outer divertor grid subset. By grid subset identifiers definition
(see ggd_subset_identifier) outer divertor is defined by index 24.
grid_ggd(1).grid_subset(4).dimension = 3grid_ggd(1).grid_subset(4).identifier.name = "outer_divertor"grid_ggd(1).grid_subset(4).identifier.index = 24grid_ggd(1).grid_subset(4).identifier.description = "Outer Divertor"
2.) Allocate 2 element AOS and for each element AOS 8 object AOS.
3.1) Defining first element - 2D cell based on 4 points each defined
using 1D spaces. The space, dimension and
index indices are used to navigate through
grid_ggd(1).space(space_index).objects_per_dimension(dimension_index).object(index):
First point:
grid_ggd(1).grid_subset(4).element(1).object(1).space = 1grid_ggd(1).grid_subset(4).element(1).object(1).dimension = 1grid_ggd(1).grid_subset(4).element(1).object(1).index = 4grid_ggd(1).grid_subset(4).element(1).object(2).space = 2grid_ggd(1).grid_subset(4).element(1).object(2).dimension = 1grid_ggd(1).grid_subset(4).element(1).object(2).index = 1
Second point:
grid_ggd(1).grid_subset(4).element(1).object(3).space = 1grid_ggd(1).grid_subset(4).element(1).object(3).dimension = 1grid_ggd(1).grid_subset(4).element(1).object(3).index = 5grid_ggd(1).grid_subset(4).element(1).object(4).space = 2grid_ggd(1).grid_subset(4).element(1).object(4).dimension = 1grid_ggd(1).grid_subset(4).element(1).object(4).index = 1
Third point:
grid_ggd(1).grid_subset(4).element(1).object(5).space = 1grid_ggd(1).grid_subset(4).element(1).object(5).dimension = 1grid_ggd(1).grid_subset(4).element(1).object(5).index = 5grid_ggd(1).grid_subset(4).element(1).object(6).space = 2grid_ggd(1).grid_subset(4).element(1).object(6).dimension = 1grid_ggd(1).grid_subset(4).element(1).object(6).index = 2
Fourth point:
grid_ggd(1).grid_subset(4).element(1).object(7).space = 1grid_ggd(1).grid_subset(4).element(1).object(7).dimension = 1grid_ggd(1).grid_subset(4).element(1).object(7).index = 4grid_ggd(1).grid_subset(4).element(1).object(8).space = 2grid_ggd(1).grid_subset(4).element(1).object(8).dimension = 1grid_ggd(1).grid_subset(4).element(1).object(8).index = 2
3.2) Defining second element - 2D cell based on 4 points each defined using 1D spaces:
First point:
grid_ggd(1).grid_subset(4).element(2).object(1).space = 1grid_ggd(1).grid_subset(4).element(2).object(1).dimension = 1grid_ggd(1).grid_subset(4).element(2).object(1).index = 4grid_ggd(1).grid_subset(4).element(2).object(2).space = 2grid_ggd(1).grid_subset(4).element(2).object(2).dimension = 1grid_ggd(1).grid_subset(4).element(2).object(2).index = 2
Second point:
grid_ggd(1).grid_subset(4).element(2).object(3).space = 1grid_ggd(1).grid_subset(4).element(2).object(3).dimension = 1grid_ggd(1).grid_subset(4).element(2).object(3).index = 5grid_ggd(1).grid_subset(4).element(2).object(4).space = 2grid_ggd(1).grid_subset(4).element(2).object(4).dimension = 1grid_ggd(1).grid_subset(4).element(2).object(4).index = 2
Third point:
grid_ggd(1).grid_subset(4).element(2).object(5).space = 1grid_ggd(1).grid_subset(4).element(2).object(5).dimension = 1grid_ggd(1).grid_subset(4).element(2).object(5).index = 5grid_ggd(1).grid_subset(4).element(2).object(6).space = 2grid_ggd(1).grid_subset(4).element(2).object(6).dimension = 1grid_ggd(1).grid_subset(4).element(2).object(6).index = 3
Fourth point:
grid_ggd(1).grid_subset(4).element(2).object(7).space = 1grid_ggd(1).grid_subset(4).element(2).object(7).dimension = 1grid_ggd(1).grid_subset(4).element(2).object(7).index = 4grid_ggd(1).grid_subset(4).element(2).object(8).space = 2grid_ggd(1).grid_subset(4).element(2).object(8).dimension = 1grid_ggd(1).grid_subset(4).element(2).object(8).index = 3

This way our grid subset outer divertor is defined.
Example 4: Unstructured grid subsets example¶
In this example we will describe unstructured grid (two time slices) in [R, Z] space consisting of
20 points (0D objects)
31 edges/lines (1D objects), and
12 surfaces/2D cells (2D objects).
This grid will contains 6 grid subsets, 3 base ones and four additional
ones, according to ggd_subset_identifier:
nodes (all nodes in the domain),
faces (all lines/edges in the domain),
cells (all surfaces/2D cells in the domain),
x_aligned_faces (all X aligned lines),
y_aligned_faces (all Y aligned lines),
outer_divertor (marked with green quadrilaterals)
Note
In this example outer_divertor grid subset won’t represent the actual outer divertor region. It is only to give an insight how to represent grid subsets in the GGD.
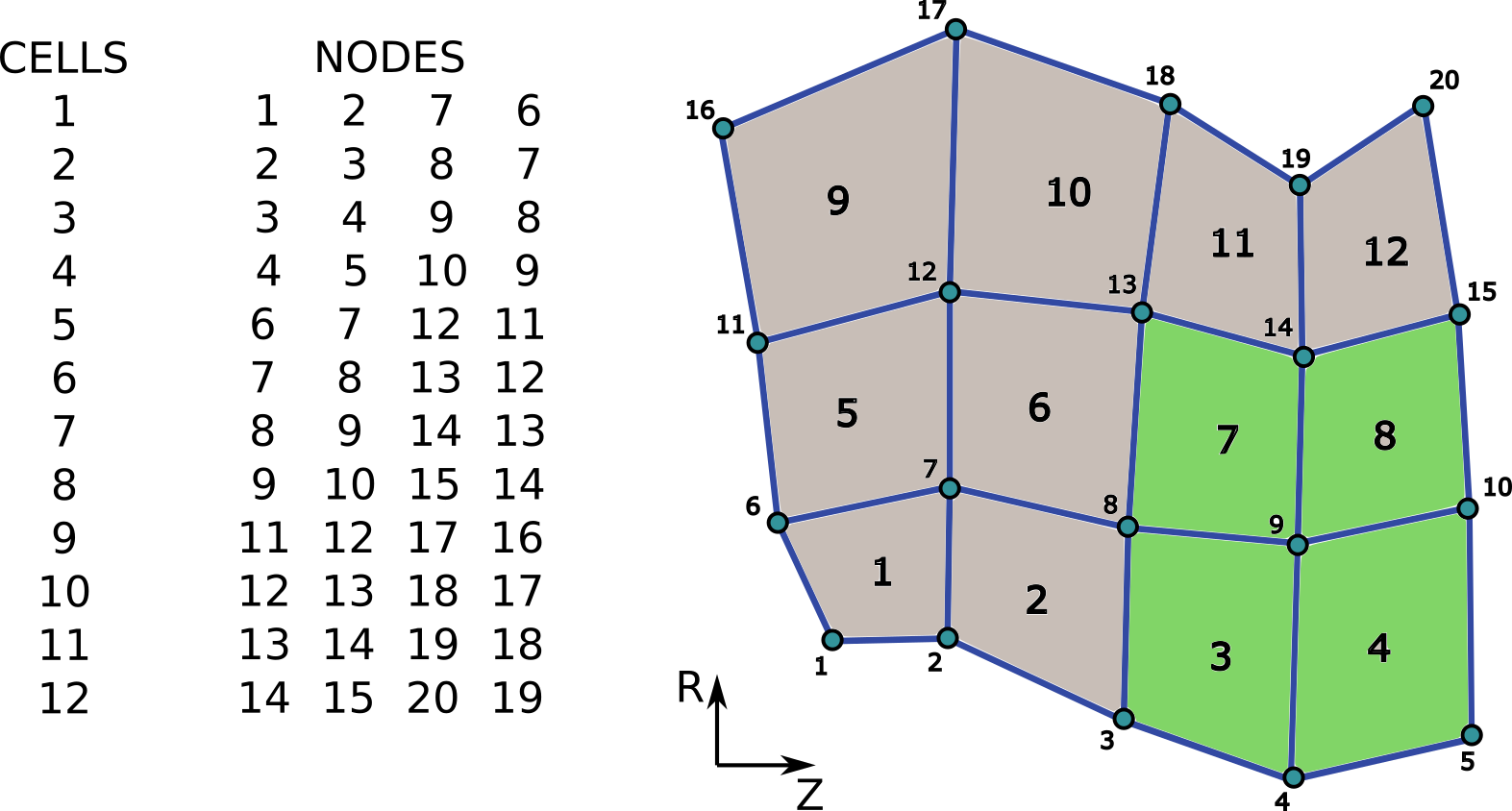
Unstructured grid used in this example and connectivity array for 2D objects - quadrilateral cells. Assumed grid subset “outer_divertor” is marked by green quadrilaterals.¶
Defining the grid¶
1.) We need to allocate one grid_ggd AOS (one AOS as we assume that grid does not change with time).
2.) In grid_ggd(1).identifier we define the grid name, index and
description:
grid_ggd(1).identifier.name = "grid_example_4"grid_ggd(1).identifier.index = 1grid_ggd(1).identifier.description = "Grid - example 4"
3.) We allocate one grid_ggd(1).space AOS.
4.) In grid_ggd(1).space(1).identifier node we define the space name,
index and description:
grid_ggd(1).space(1).identifier.name = "space_example_4"grid_ggd(1).space(1).identifier.index = 1grid_ggd(1).space(1).identifier.description = "Space - example 4"
5.) In grid_ggd(1).space(1).geometry_type.index leaf we define
standard geometry -> index 0
grid_ggd(1).space(1).geometry_type.index = 0
6.) In grid_ggd(1).space(1).coordinates_type leaf we define two
coordinates following the coordinate_identifier this way defining 2D
space. We define 4 and 3 meaning R and Z coordinates
according to coordinate_identifier:
grid_ggd(1).space(1).coordinate_types = [4, 3]
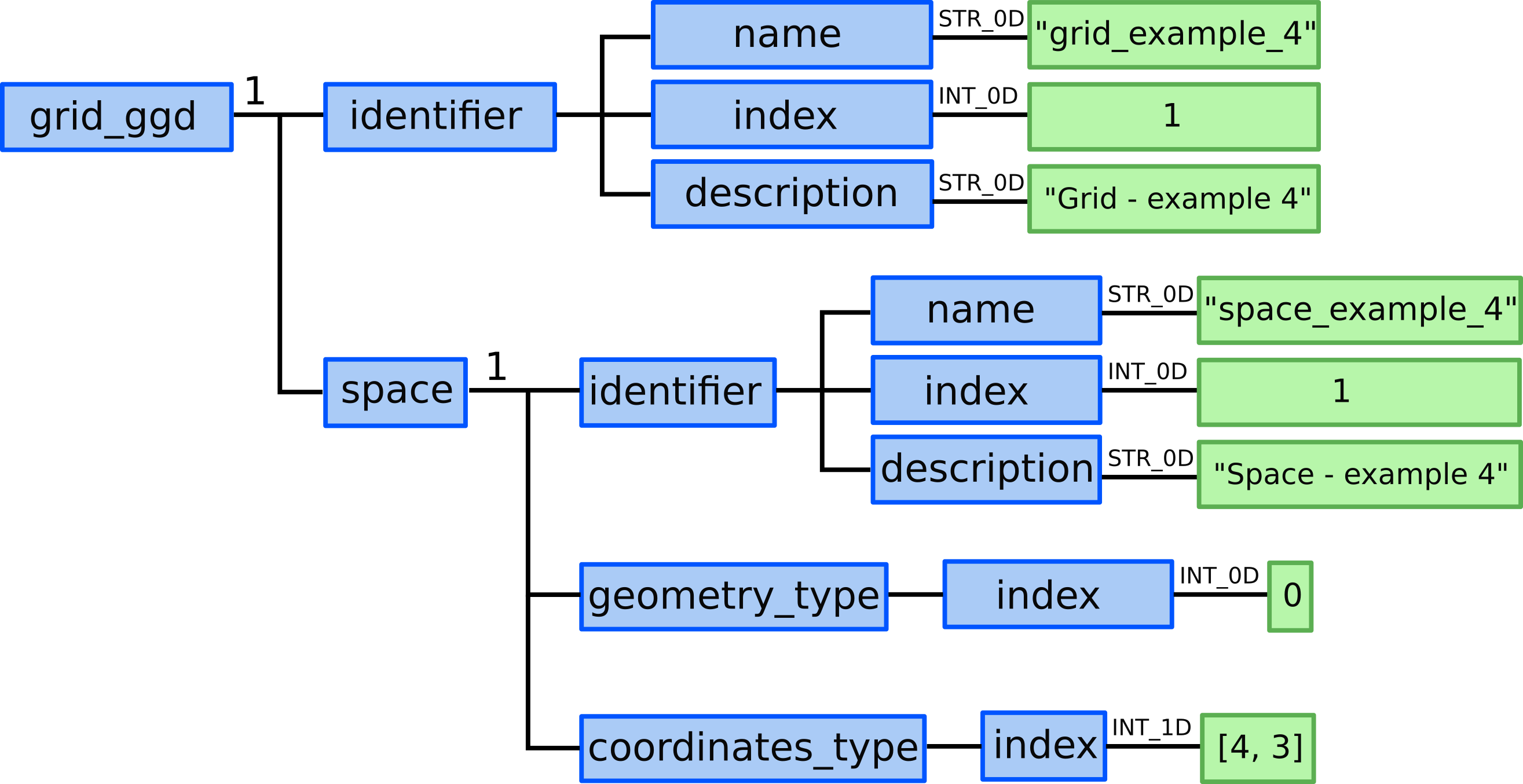
Representation of defining grid and space identifier name, index, and description; and space geometry_type and coordinates_type in GGD.¶
7.) We allocate three grid_ggd(1).space(1).objects_per_dimension AOSs:
for 0D objects - points, 1D objects - edges/lines, and
2D objects - surfaces/2D cells.
8.1) We allocate 20 ...objects_per_dimension(1).object AOS as our grid
consists of 20 points.
8.2) We define 20 points by defining their geometry (note that the coordinates are simplified and marked by \({r}\) and \({z}\) symbols instead of values according to the example):
...objects_per_dimension(1).object(1).geometry = [r_1, z_1]...objects_per_dimension(1).object(2).geometry = [r_2, z_2]...objects_per_dimension(1).object(3).geometry = [r_3, z_3]…
...objects_per_dimension(1).object(20).geometry = [r_20, z_20]
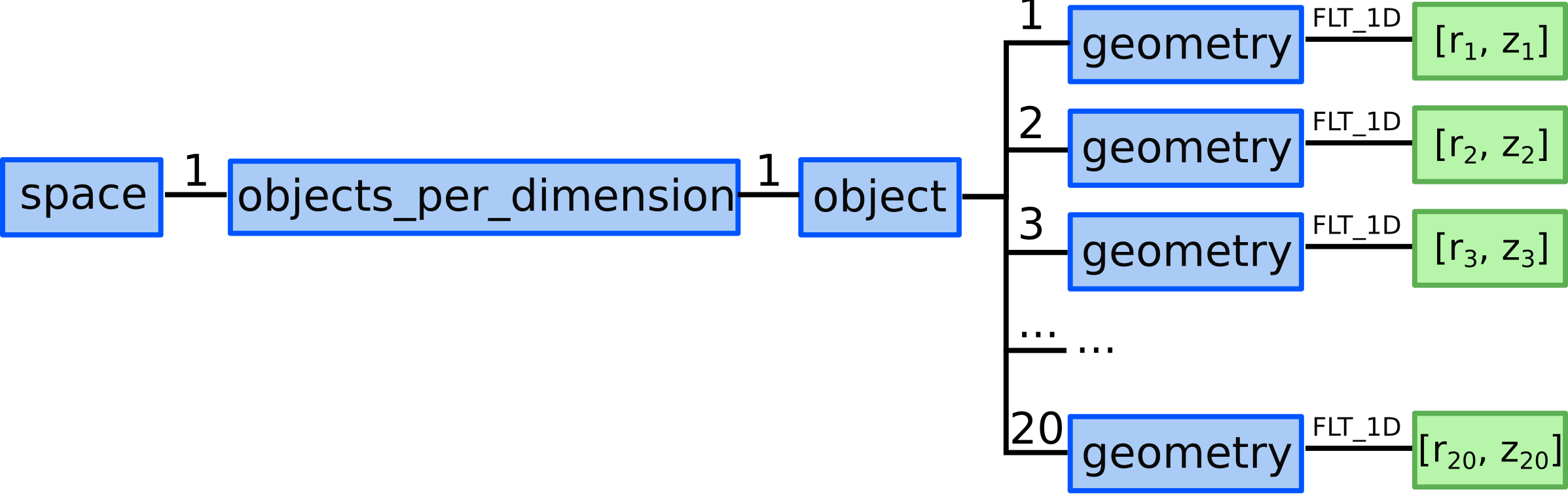
Representation of defining all 0D objects/points/nodes in the domain with GGD.¶
9.1) We allocate 31 ...objects_per_dimension(2).object AOS as our grid
consists of 31 lines.
9.2) We define 31 lines by defining nodes that compose each line. First we define the “x-aligned (r-aligned)” lines and then the “y-aligned (z-aligned” lines.
...objects_per_dimension(2).object(1).nodes = [1, 2]...objects_per_dimension(2).object(2).nodes = [2, 3]...objects_per_dimension(2).object(3).nodes = [3, 4]...objects_per_dimension(2).object(4).nodes = [4, 5]...objects_per_dimension(2).object(5).nodes = [6, 7]...objects_per_dimension(2).object(6).nodes = [7, 8]...objects_per_dimension(2).object(7).nodes = [8, 9]...objects_per_dimension(2).object(8).nodes = [9, 10]...objects_per_dimension(2).object(9).nodes = [11, 12]...objects_per_dimension(2).object(10).nodes = [12, 13]...objects_per_dimension(2).object(11).nodes = [13, 14]...objects_per_dimension(2).object(12).nodes = [14, 15]...objects_per_dimension(2).object(13).nodes = [16, 17]...objects_per_dimension(2).object(14).nodes = [17, 18]...objects_per_dimension(2).object(15).nodes = [18, 19]...objects_per_dimension(2).object(16).nodes = [19, 20]...objects_per_dimension(2).object(17).nodes = [1, 6]...objects_per_dimension(2).object(18).nodes = [6, 11]...objects_per_dimension(2).object(19).nodes = [11, 16]...objects_per_dimension(2).object(20).nodes = [2, 7]...objects_per_dimension(2).object(21).nodes = [7, 12]...objects_per_dimension(2).object(22).nodes = [12, 17]...objects_per_dimension(2).object(23).nodes = [3, 8]...objects_per_dimension(2).object(24).nodes = [8, 13]...objects_per_dimension(2).object(25).nodes = [13, 18]...objects_per_dimension(2).object(26).nodes = [4, 9]...objects_per_dimension(2).object(27).nodes = [9, 14]...objects_per_dimension(2).object(28).nodes = [14, 19]...objects_per_dimension(2).object(29).nodes = [5, 10]...objects_per_dimension(2).object(30).nodes = [10, 15]...objects_per_dimension(2).object(31).nodes = [15, 20]
Note
The ...objects_per_dimension(2).object(:).measure could be filled
with the line length, however we don’t need that information here.

Representation of defining all 1D objects/lines in the domain with GGD.¶
10.1) We allocate 12 ...objects_per_dimension(3).object AOS as our grid
consists of 12 quadrilateral cells.
10.2) We define connectivity array for 2D objects - 12 quads by defining nodes that compose each quad element.
...objects_per_dimension(3).object(1).nodes = [1, 2, 7, 6]...objects_per_dimension(3).object(2).nodes = [2, 3, 8, 7]...objects_per_dimension(3).object(3).nodes = [3, 4, 9, 8]...objects_per_dimension(3).object(4).nodes = [4, 5, 10, 9]...objects_per_dimension(3).object(5).nodes = [6, 7, 12, 11]...objects_per_dimension(3).object(6).nodes = [7, 8, 13, 12]...objects_per_dimension(3).object(7).nodes = [8, 9, 14, 13]...objects_per_dimension(3).object(8).nodes = [9, 10, 15, 14]...objects_per_dimension(3).object(9).nodes = [11, 12, 17, 16]...objects_per_dimension(3).object(10).nodes = [12, 13, 18, 17]...objects_per_dimension(3).object(11).nodes = [13, 14, 19, 18]...objects_per_dimension(3).object(12).nodes = [14, 15, 20, 19]
Note
The ...objects_per_dimension(3).object(:).measure could be filled
with the quad surface area, however we don’t need that information
here.
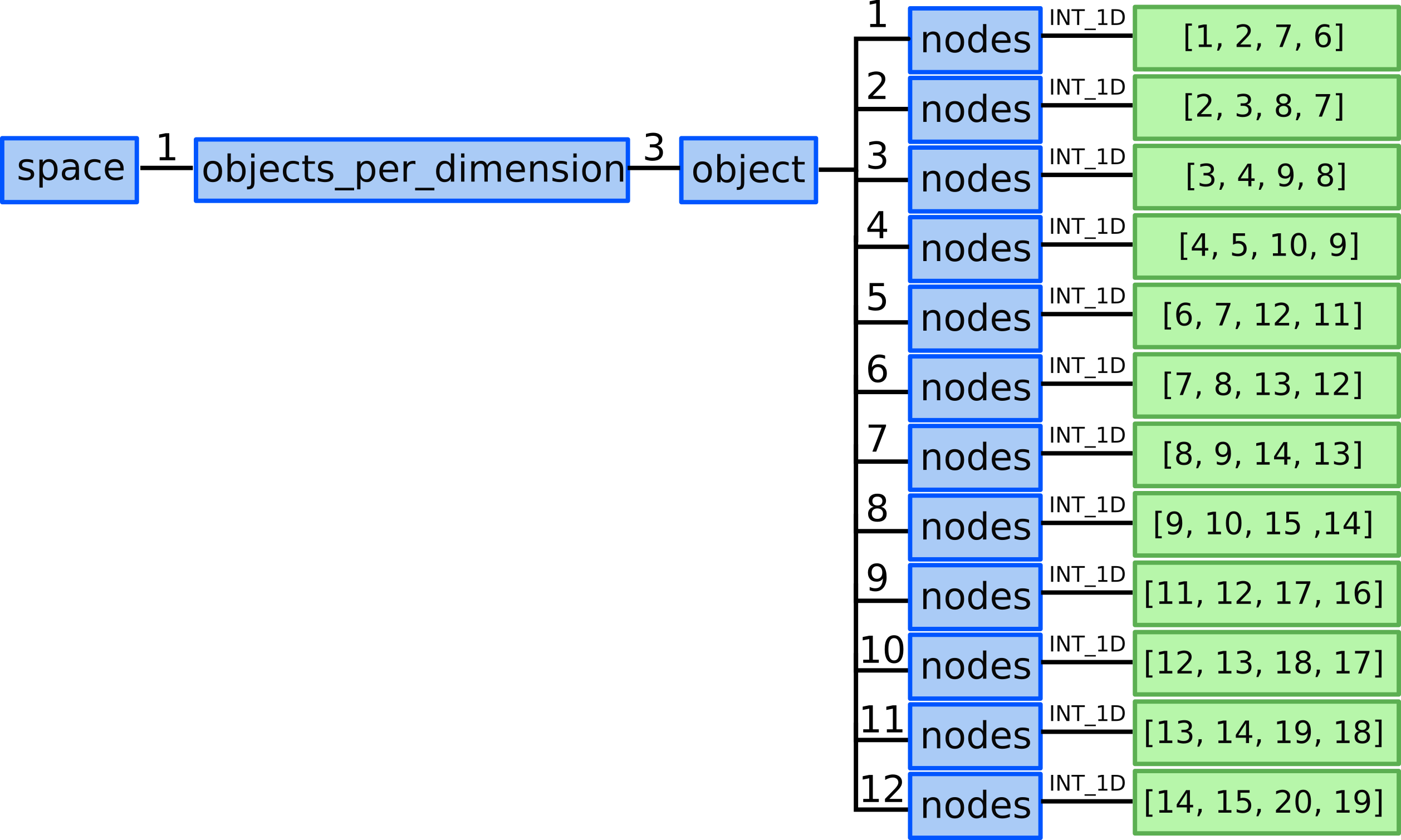
Representation of defining all 2D objects/quads in the domain with GGD.¶
Defining grid subsets¶
First we need to define the grid subsets correlating to the 0D, 1D and 2D objects in the domain. This is due to Data Dictionary description, as physical quantities (data fields) correlate directly to grid subsets and not to the grid itself.
Second, we will define 3 more grid subsets: x_aligned_faces, y_aligned_faces, and our “outer divertor” grid subset, giving us a total of 6 grid subsets.
The agreed grid subset labels can be found here:
ggd_subset_identifier.
Allocate 6
grid_ggd(1).grid_subsetAOSs:
12.1) Define nodes grid subset. By definition nodes grid subset is composed by all 0D objects defined in this grid_ggd AOS so the element AOS can be left empty.
grid_ggd(1).grid_subset(1).dimension = 1grid_ggd(1).grid_subset(1).identifier.name = "nodes"grid_ggd(1).grid_subset(1).identifier.index = 1grid_ggd(1).grid_subset(1).identifier.description = "All nodes in the domain"
12.2) Define faces grid subset. By definition faces grid subset is composed by all 0D objects defined in this grid_ggd AOS so the element AOS can be left empty.
grid_ggd(1).grid_subset(2).dimension = 2grid_ggd(1).grid_subset(2).identifier.name = "faces"grid_ggd(1).grid_subset(2).identifier.index = 2grid_ggd(1).grid_subset(2).identifier.description = "All lines in the domain"
12.3) Define cells grid subset. By definition cells grid subset is composed by all 0D objects defined in this grid_ggd AOS so the element AOS can be left empty.
grid_ggd(1).grid_subset(3).dimension = 3grid_ggd(1).grid_subset(3).identifier.name = "cells"grid_ggd(1).grid_subset(3).identifier.index = 3grid_ggd(1).grid_subset(3).identifier.description = "All 2D cells in the domain"

Defining the three base grid subsets - nodes, faces and cells.¶
12.3) Define x_alignes_faces grid subset. Here we need to define
16 elements each composed by single 1D object - face. Other option could
be to define 16 elements each composed by two 0D objects - points, however
we already have the lines defined in objects_per_dimension(2) so this is
not necessary.
grid_ggd(1).grid_subset(4).dimension = 2grid_ggd(1).grid_subset(4).identifier.name = "x_aligned_faces"grid_ggd(1).grid_subset(4).identifier.index = 3grid_ggd(1).grid_subset(4).identifier.description = "X aligned faces"grid_ggd(1).grid_subset(4).element(1).object(1).space = 1grid_ggd(1).grid_subset(4).element(1).object(1).dimension = 2grid_ggd(1).grid_subset(4).element(1).object(1).index = 1grid_ggd(1).grid_subset(4).element(2).object(1).space = 1grid_ggd(1).grid_subset(4).element(2).object(1).dimension = 2grid_ggd(1).grid_subset(4).element(2).object(1).index = 2
In this case all object AOSs hold the same space and dimension, only
the index is different, navigating to different object in the same space
under the same dimension. Following that, only the different index leaf
contents will be shown below.
grid_ggd(1).grid_subset(4).element(3).object(1).index = 3grid_ggd(1).grid_subset(4).element(4).object(1).index = 4grid_ggd(1).grid_subset(4).element(5).object(1).index = 5grid_ggd(1).grid_subset(4).element(6).object(1).index = 6grid_ggd(1).grid_subset(4).element(7).object(1).index = 7grid_ggd(1).grid_subset(4).element(8).object(1).index = 8grid_ggd(1).grid_subset(4).element(9).object(1).index = 9grid_ggd(1).grid_subset(4).element(10).object(1).index = 10grid_ggd(1).grid_subset(4).element(11).object(1).index = 11grid_ggd(1).grid_subset(4).element(12).object(1).index = 12grid_ggd(1).grid_subset(4).element(13).object(1).index = 13grid_ggd(1).grid_subset(4).element(14).object(1).index = 14grid_ggd(1).grid_subset(4).element(15).object(1).index = 15grid_ggd(1).grid_subset(4).element(16).object(1).index = 16

Defining the x_aligned_faces grid subset in GGD.¶
12.4) Define y_alignes_faces grid subset. Here we need to define 15 elements each composed by single 1D object - face.
grid_ggd(1).grid_subset(5).dimension = 2grid_ggd(1).grid_subset(5).identifier.name = "y_aligned_faces"grid_ggd(1).grid_subset(5).identifier.index = 4grid_ggd(1).grid_subset(5).identifier.description = "Y aligned faces"grid_ggd(1).grid_subset(5).element(1).object(1).space = 1grid_ggd(1).grid_subset(5).element(1).object(1).dimension = 2grid_ggd(1).grid_subset(5).element(1).object(1).index = 17grid_ggd(1).grid_subset(5).element(2).object(1).space = 1grid_ggd(1).grid_subset(5).element(2).object(1).dimension = 2grid_ggd(1).grid_subset(5).element(2).object(1).index = 18
In this case all object AOSs hold the same space and dimension, only
the index is different, navigating to different object in the same space
under the same dimension. Following that, only the different index leaf
contents will be shown below.
grid_ggd(1).grid_subset(5).element(3).object(1).index = 19grid_ggd(1).grid_subset(5).element(4).object(1).index = 20grid_ggd(1).grid_subset(5).element(5).object(1).index = 21grid_ggd(1).grid_subset(5).element(6).object(1).index = 22grid_ggd(1).grid_subset(5).element(7).object(1).index = 23grid_ggd(1).grid_subset(5).element(8).object(1).index = 24grid_ggd(1).grid_subset(5).element(9).object(1).index = 25grid_ggd(1).grid_subset(5).element(10).object(1).index = 26grid_ggd(1).grid_subset(5).element(11).object(1).index = 27grid_ggd(1).grid_subset(5).element(12).object(1).index = 28grid_ggd(1).grid_subset(5).element(13).object(1).index = 29grid_ggd(1).grid_subset(5).element(14).object(1).index = 30grid_ggd(1).grid_subset(5).element(15).object(1).index = 31
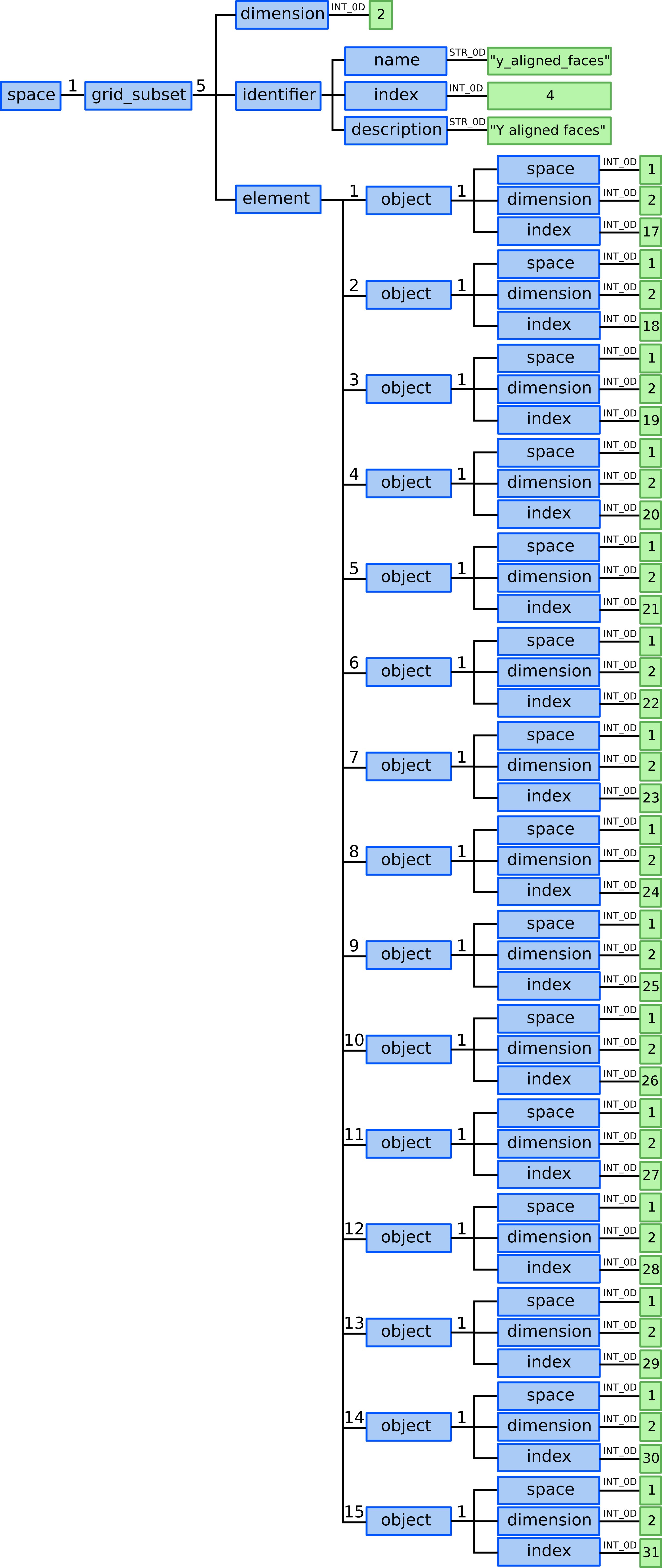
Defining the y_aligned_faces grid subset in GGD.¶
12.5) Define our “outer_divertor” grid subset. Here we need to define 12 elements each composed by single 2D object - cell/quad.
grid_ggd(1).grid_subset(6).dimension = 3grid_ggd(1).grid_subset(6).identifier.name = "outer_divertor"grid_ggd(1).grid_subset(6).identifier.index = 24grid_ggd(1).grid_subset(6).identifier.description = "Cells defining the outer divertor region"grid_ggd(1).grid_subset(6).element(1).object(1).space = 1grid_ggd(1).grid_subset(6).element(1).object(1).dimension = 2grid_ggd(1).grid_subset(6).element(1).object(1).index = 1grid_ggd(1).grid_subset(6).element(2).object(1).space = 1grid_ggd(1).grid_subset(6).element(2).object(1).dimension = 2grid_ggd(1).grid_subset(6).element(2).object(1).index = 2grid_ggd(1).grid_subset(6).element(2).object(1).space = 1grid_ggd(1).grid_subset(6).element(2).object(1).dimension = 2grid_ggd(1).grid_subset(6).element(3).object(1).index = 3grid_ggd(1).grid_subset(6).element(2).object(1).space = 1grid_ggd(1).grid_subset(6).element(2).object(1).dimension = 2grid_ggd(1).grid_subset(6).element(4).object(1).index = 4
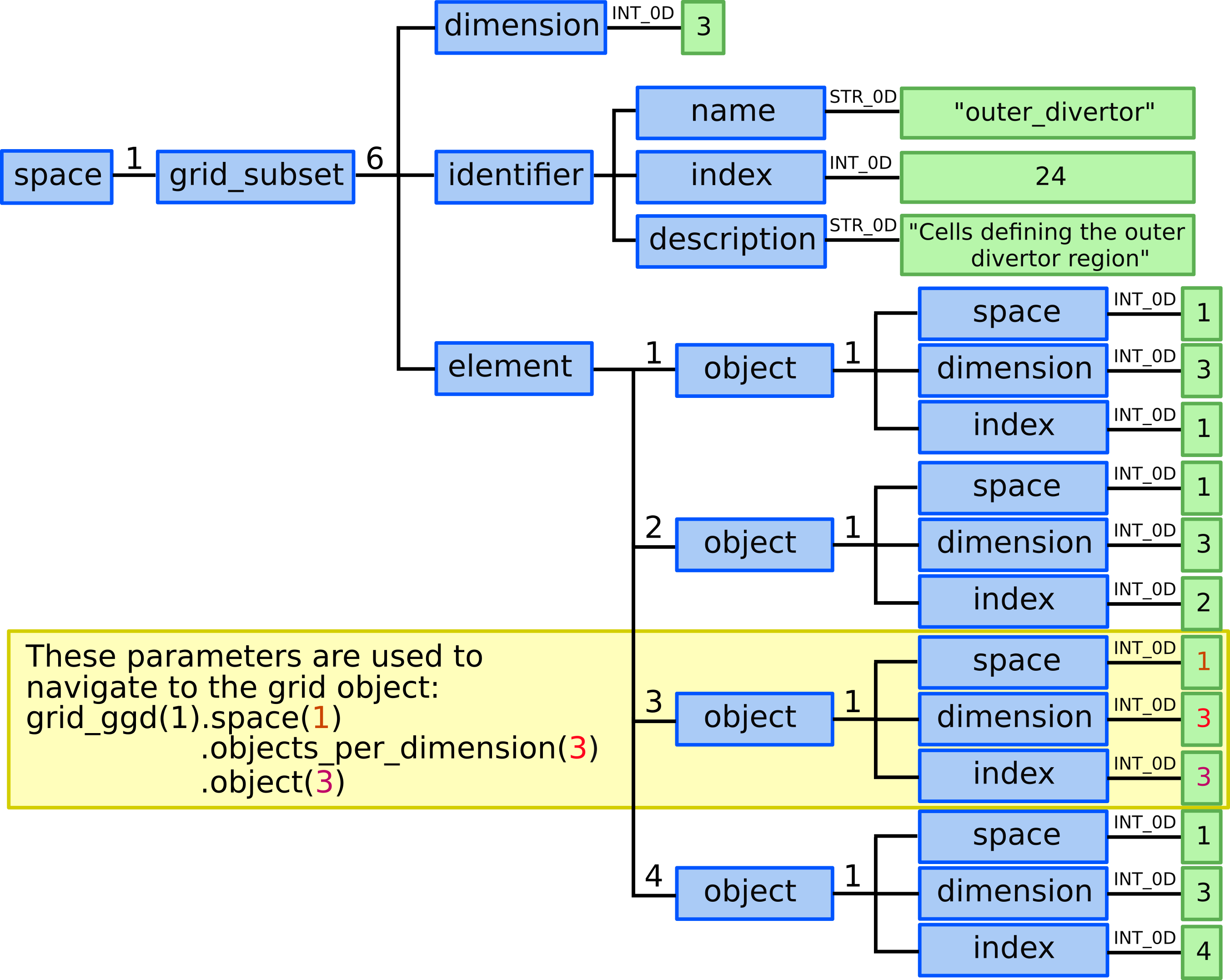
Defining the outer_divertor grid subset in GGD.¶
Defining physical quantities¶
In this example we assume that we have electron temperature
quantities/values for two time slices (the grid is static) at times
1.0ms and 2.0ms that correlate to our 20 points and 12 quads. Defining them in GGD is done in the next
way:
13.) We need to allocate two ggd AOS (two time slices).
14.) We specify time value for both time slices:
ggd(1).time= 1.0ggd(2).time= 2.0
Electron temperature¶
15.) We allocate three ggd(1).electrons.temperature AOSs as in this case
we have available (we assume) electron temperature quantities for three
grid subsets - nodes, cells and outer_divertor.
First time slice¶
16.) In ggd(1).electrons.temperature(1) we define grid_index,
grid_subset_index and values leafs.
grid_index and grid_subset_index are used to navigate
to grid which has grid_ggd(:).identifier.index == grid_index
and :grid_ggd(:).grid_subset(:).identifier.index == grid_subset_index.
In this case it would navigate to our nodes grid subset.
ggd(1).electrons.temperature(1).grid_index= 1ggd(1).electrons.temperature(1).grid_subset_index= 1ggd(1).electrons.temperature(1).values= e.g. [1.1, 2.2, …, 20.20] (one value per point, 20 values)
17.) In ggd(1).electrons.temperature(2) we define grid_index,
grid_subset_index and values leafs.
In this case it would navigate to our cells grid subset.
ggd(1).electrons.temperature(2).grid_index= 1ggd(1).electrons.temperature(2).grid_subset_index= 5ggd(1).electrons.temperature(2).values= e.g. [1.1, 2.2, …, 12.12] (one value per quad, 12 values)
18.) In ggd(1).electrons.temperature(3) we define grid_index,
grid_subset_index and values leafs.
In this case it would navigate to our outer_divertor grid subset.
ggd(1).electrons.temperature(3).grid_index= 1ggd(1).electrons.temperature(3).grid_subset_index= 24ggd(1).electrons.temperature(3).values= e.g. [3.3, 4.4, 7.7, 8.8] (one value per quad, 4 values)
Second time slice¶
19.) In ggd(2).electrons.temperature(1) we define grid_index,
grid_subset_index and values leafs.
In this case it would navigate to our nodes grid subset.
ggd(2).electrons.temperature(1).grid_index= 1ggd(2).electrons.temperature(1).grid_subset_index= 1ggd(2).electrons.temperature(1).values= e.g. [2.2, 3.3, …, 21.21] (one value per point, 20 values)
20.) In ggd(2).electrons.temperature(2) we define grid_index,
grid_subset_index and values leafs.
In this case it would navigate to our cells grid subset.
ggd(2).electrons.temperature(2).grid_index= 1ggd(2).electrons.temperature(2).grid_subset_index= 5ggd(2).electrons.temperature(2).values= e.g. [2.2, 3.3, …, 13.13] (one value per quad, 12 values)
21.) In ggd(2).electrons.temperature(3) we define grid_index,
grid_subset_index and values leafs.
In this case it would navigate to our outer_divertor grid subset.
ggd(2).electrons.temperature(3).grid_index= 1ggd(2).electrons.temperature(3).grid_subset_index= 24ggd(2).electrons.temperature(3).values= e.g. [4.4, 5.5, 8.8, 9.9] (one value per quad, 4 values)
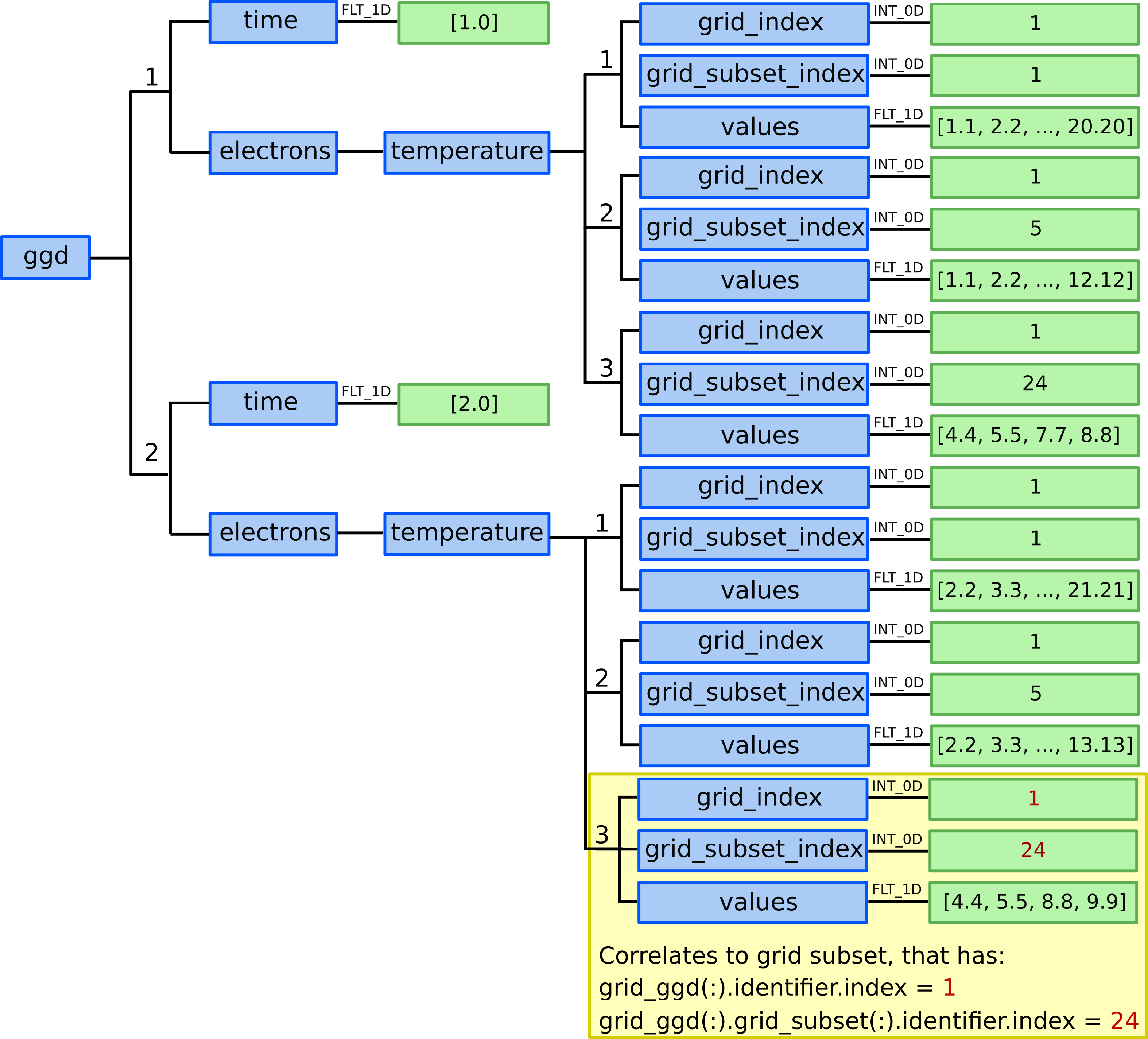
Defining electron temperature data fields for two time slices and three grid subsets - nodes, cells, and our outer_divertor.¶
MHD example: Combined unstructured and Fourier space¶
The following example is based on the [JOREK] non-linear MHD code.
JOREK uses Bezier FEM description in poloidal plane and Fourier
interpolation in toroidal direction. So far, the implementaion of such
geometry has not been treated in similar codes. The example follows
jorek/util/IMAS/JOREK2IDS/jorekjorekHDF5toIDS.py python script
that reads jorek HDF5 files and writes IDS. The most important
properties that are stored are 2D grid, it’s properties and values.
The discretization of JOREK is well described in the paper [Czarny-Huysmans-2008].
For all variables indexed by \(\nu=1\dots N_{var}\) the distibution \(X(s,t,\varphi)\) of quantity \(X\) at the toroidal position \(\varphi\) in a given finite element can be expressed by
where i=element.vertex(k) is the global node index corresponding to
the vertex k of the finite element. Here, \(N_{var}\) denotes the
number of physical variables, \(s\) and \(t\) are the poloidal
local coordinates, H are Bezier basis functions (polynomials),
element.size are geometrical coefficients for the element,
\(N_{vert}=4\) the number of vertices in each
element, \(N_{ord}=4\) the number of degrees of freedom per
vertex, and \(N_{tor}\) the number of different toroidal Fourier
modes. The quantity \(Z_l(\varphi)\) corresponds to the value of
the \(l\)-th Fourier mode at the toroidal position \(\varphi\).
The table below lists which Fourier modes correspond to the different
mode indices \(l\) with \(n_p\) as peridicity of tyhe simulation
\(l\) |
1 |
2 |
3 |
4 |
5 |
… |
|---|---|---|---|---|---|---|
\(Z_l(\varphi)\) |
1 |
\(\cos(n_p\varphi_p)\) |
\(\sin(n_p\varphi_p)\) |
\(\cos(2n_p\varphi)\) |
\(\sin(2n_p\varphi)\) |
… |
Fourier Periodicity - \(n_p\)¶
- Type of space geometry (
grid_ggd.space.geometry_type) 0: standard,
1:Fourier,
>1: Fourier with periodicity
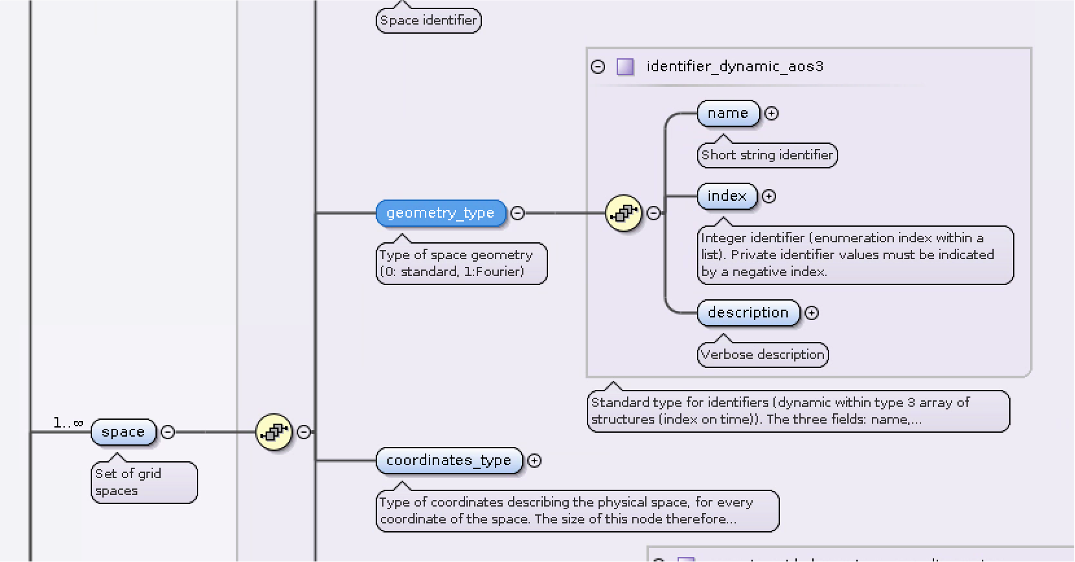
Defining the grid¶
Note
If possible, we should always decompose the discretisation into spaces: independent discretisation of individual directions.
Following above rule and JOREK discretisation there are two grid_ggd spaces:
Space 1 is two-dimensional (R,Z) space of unstructured grid with geometry_2d associated to nodes and cells. Note that the R, Z coordinates are expressed as every other quantity \(X(s,t,\varphi)\). For tokamak applications, the (R,Z) grid does not vary toroidally and only the n=0 harmonic is employed. In this case and using JOREK notation, R and Z are expressed as
\(R(s,t) =\sum_{k=1}^{N_{vert}}\sum_{j=1}^{N_{ord}}\texttt{nodes(i).x(j,1)} \cdot\texttt{H}(k,j;s,t) \cdot\texttt{element.size(k,j)}\) \(Z(s,t) =\sum_{k=1}^{N_{vert}}\sum_{j=1}^{N_{ord}}\texttt{nodes(i).x(j,2)} \cdot\texttt{H}(k,j;s,t) \cdot\texttt{element.size(k,j)}\)
The geometrical coefficients representing the grid (
x) are stored asobjects_per_dimension[0].object[i].geometry_2d(j,:)=nodes(i).x(j,:)And the elements sizes \(1, d_{uk}, d_{vk}, d_{uv}d_{vk}\) under
objects_per_dimension[2].object[i_elm].geometry_2d(:,:)=element.size(:,:)
Space 2 is one-dimensional Fourier space with \(\varphi\) and
geometry_type.index >= 1(Fourier) with geometry objects (1, 2, 3, …, number of harmonics)
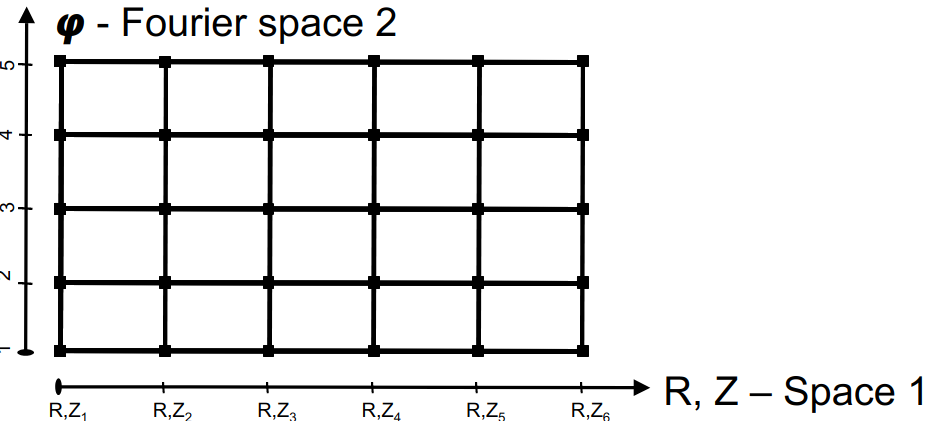
Firstly we write every node and every cell in
grid_ggd/space/object_per_dimension. Nodes are stored under index
“0”. Each node has \(R, Z\) coordinate in .../geometry with
derivatives (in R and Z direction and mixed) needed for
calculating control points and grid’s curvature in
.../geometry_2d. Cells are written in similar manner with their
connectivity in geometry and 2D array size in
geometry_2d. Size describes distances from nodes to cell’s control
points which is also necessary for plotting more precise grid. Both
size and derivatives are explained in detail in thesis by Daan Van
Vugt [VanVugt19].
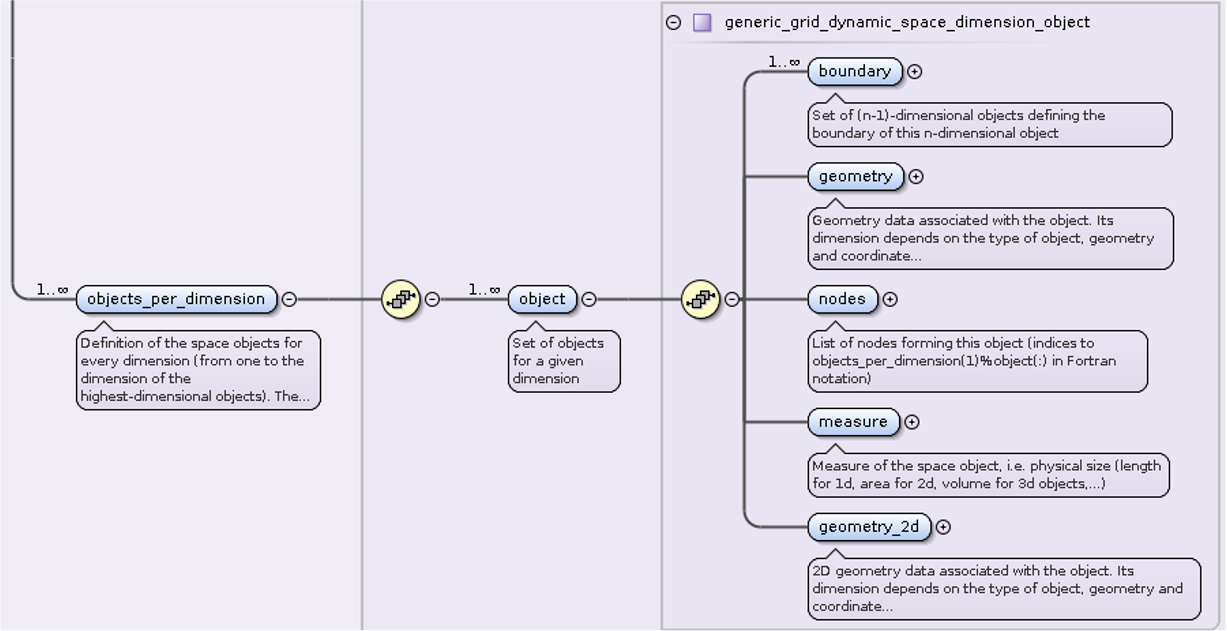
Defining physical quantities¶
For each physical quantity (temperature, psi, j..) values and it’s
derivatives must be stored for each node. Values are in
grid/variable/values
Values (Te, n, w, …), stored under GGD, with the above two spaces form a “structured” (implicitly defined) grid of node values, where explicitly RZ values of first harmonics are saved first, then RZ values of the second harmonics follow up to the last RZ harmonics. Similarly, coefficients on the nodes are saved. This definition follows column major (FORTRAN) notation, meaning that with varying first index (R) the values are close together in the memory and that the last index (\(\varphi\) in Fourier space) defines RZ block of values in memory. At the moment, the correspondance between \(X(s,t,\varphi)\) and GGD is, for example for the toroidal current density
mhd_ids.ggd[islice].j_tor[0].coefficients[l,m(i,j) ]=nodes(i).values(l,j,index_jtor)
and m(i,j)=i+(j-1)N_dof.
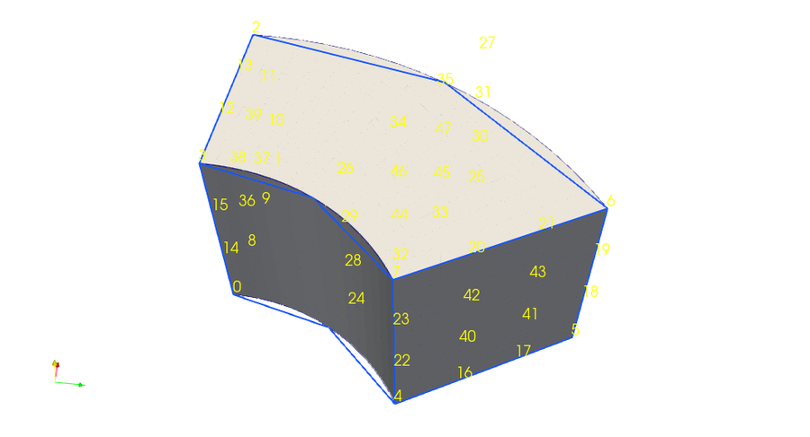
Bezier elements in VTK¶
Bezier geometry is the most complex, but it enables us to increase nonlinear subdivision level for same number of points within visualisation platform (such as paraview), because it uses Bezier polynomials to interpolate more points from initial control points. Calculation of control points in RZ plane used here is explained in thesis [VanVugt19].
For 2D these control points are used to create
Bezier quadrilaterals. As
a result each cell is made of 4×4 nodes. When we move to 3D, Bezier
hexahedrons are
made of 3 planes of 4×4 points, which gives us 48 nodes per
cell. Because our cell is not 4x4x4 we just skip nodes that are not
there. Anisotropic structure of cell’s shape is specified by
SetHigherOrderDegrees() with shape[3,3,2] in
jorek/utils/jorek_read_h5.py. (Even though cells are made of
4x4x3 points, one number less must be given for each dimension.)
Therefore, we have anisotropic Bezier element with:
In poloidal plane exact Bezier cubic non-rational quads
In toroidal direction an approximation between toroidal planes with rational quadratic Bezier curves.
For sake of clarity a jorek/util/IMAS/JOREK2IDS/IDS_to_VTK.py
script is made to read previously written data and makes .vtu
for visualisation with ParaView 5.9.1 or higher. Once again we must
choose some arguments to specify what we want our plot to look
like. The most important are “number of planes” and
\(\varphi\). Script writes object with Bezier cells if not decided
differently. If Bezier is disabled the plane quadrilateral mesh (2D)
without Fourier harmonics is used. However, node numbering in this
case corresponds to JOREK node and cell numbering (without
subdivision).